MINDy
Contents
MINDy
The MINDy algorithm (Modulator Inference by Network Dynamics) uses gene expression data to determine whether a putative modulator gene (Mj) influences the regulatory activity of a transcription factor gene (TF) over a set of target genes (Ti). This influence is measured in terms of whether there is a change in the correlation (measured as mutual information) of expression between the TF and its targets Ti conditional on a change in the expression of Mj. The mutual information values used in MINDy are calculated using the ARACNe algorithm, which is also a part of geWorkbench.
Outline of MINDy calculations
- A microarray gene expression dataset is selected.
- The user specifies a set of one or more candidate modulator genes (Mj), a hub transcription factor (TF), and a set of putative targets of the transcription factor (Ti).
- Parameters for the MINDy run are set.
- Using the expression value of the chosen modulator gene Mj, the arrays in the experiment are ordered (as columns in the data matrix), from lowest to highest.
- Two subsets of arrays are then chosen from each end (tail) of the ordered list. One subset contains arrays in which Mj shows the lowest expression, and the other subset contains arrays in which Mj shows the highest expression. The subsets are non-overlapping. A typical trial might involve assigning the lowest 35% of the arrays to the low group (M-), as measured by expression of Mj, and the highest 35% to the high group (M+). The remaining arrays are not further considered.
- For each target Ti, the conditional mutual information between the hub TF and the target is then calculated for the array subsets M+ and M- separately, and the difference is taken (delta (MI)).
- The resulting delta (MI)s are displayed. At present, a p-value is not calculated on the delta (MI). Larger values of delta (I) may indicate an interesting change in the mutual information conditional on the expression of the modulator, that is, the modulator has an effect on the correlation of expression between the hub TF and the target gene.
- The sign of the influence of the modulator is also displayed, e.g. does increasing the expression of the modulator gene Mj increase (or decrease) the correlation of expression between the hub TF and the target gene?
Important notes on the calculation
MI Thresholds
The unconditional mutual information calculation is intended to be used in calculating a significance value on the final delta MI score. This feature is not yet implemented. The conditional MI score set will influence how many target markers are returned - the lower the threshold, the more targets will be returned. A target has to have a value above the set threshold in at least one of the two conditional ARACNe runs in order to be included in the output data. The threshold should be kept as close to zero as practical to avoid truncation effects on sub-threshold values.
delta (MI) Threshold
In the current release of geWorkbench (v1.6.3), the significance of the delta (MI) value is not calculated. This p-value requires a significant post-processing calculation that is planned for inclusion with the release of MINDy2 in geWorkbench (Spring 2009).
Prerequisites for MINDy calculations
The MINDy calculation contains certain assumptions (Wang et al, unpublished):
(a) the expression of the modulator (gm)must have a sufficient expression range to separate its two expression tails compared to the experimental noise level. This can be done by running the deviation filter (Filtering component) on the dataset before starting the MINDy calculation.
(b) Any modulator whose expression profile (Mj) is not statistically independent of that of the hub transcription factor (TF) must be excluded. This can be determined using a mutual information calculation (ARACNE). This functionality is not currently directly implemented within MINDy in geWorkbench.
(c) Optimally, at least 100 separate microarrays should be included in the analysis, with a range of different expression conditions (distinct cellular phenotypes).
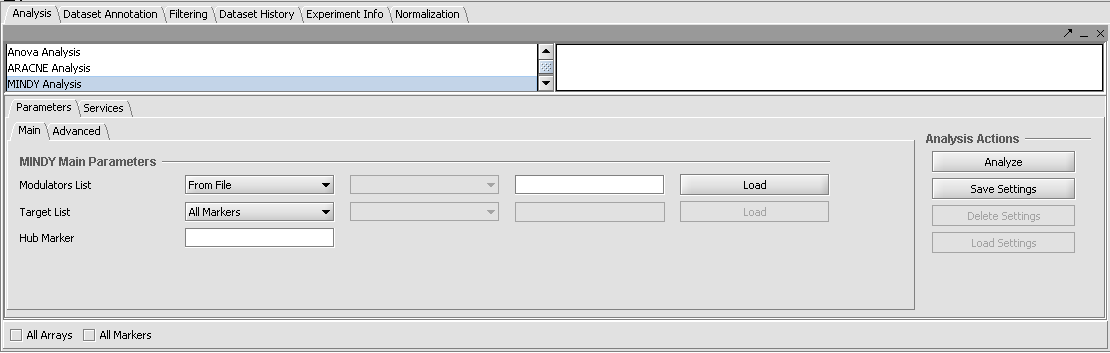
Setting the Main Parameters
Modulators List - [From File or From Set] - The list of candidate modulators can either be loaded from a file as a comma separated list, or a set of markers can be selected from the Markers component. The gene expression profiles of the modulators should be independent of that of the hub TF gene as measured by mutual information. This could be determined using a preliminary run of ARACNE including just the modulators and the transcription factor.
NOTE- in the next version of MINDy, the ability to calculate a p-value on the conditional mutual information score will be implemented. When available, it will be useful to limit the number of modulator genes tested to minimize the multiple testing correction.
Hub Marker - Enter the marker ID for a known or putative transcription factor gene. The Hub marker can be entered directly in the text field, or the most recently selected marker in the Markers component will be used. Note that even if one directly types in a marker name, it will be replaced if any selection is made in the Markers component, either in the list or in the default Marker set "Selection".
Target List - [All Markers, From File, or From Set] - The target list should be composed of genes thought to be regulated by the Hub Marker transcription factor. The list of target markers can be loaded from a file containing a comma separated list, or a set of markers can be selected from the Markers component. Alternatively, All Markers can be selected.
(Note - the "All Markers" checkbox at the bottom of the Analysis component should not be used in the MINDy component).
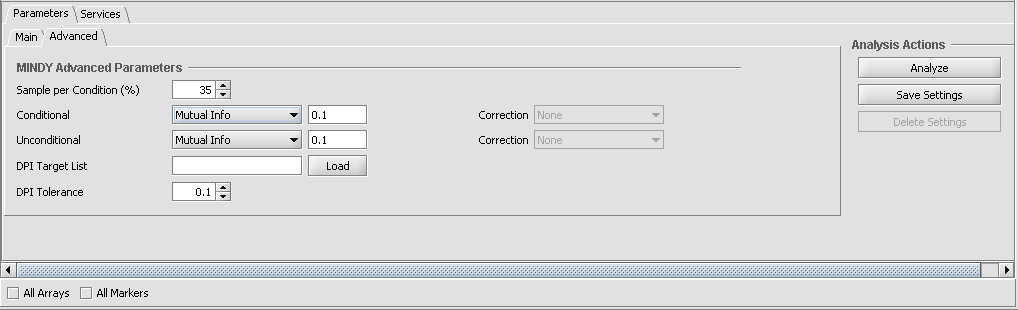
Setting the Advanced Parameters
Sample per Condition (%) - MINDy calculates the difference in mutual information for the TF-Target interaction between the set where the modulator gene is most expressed (+) and the set where the modulator gene is least expressed (-). This parameter specifies the percentage of the available samples to include in each group. E.g. 35% means that the top and bottom 35% of a list of samples ranked by expression would be used.
Unconditional and Conditional- The underlying ARACNe calculation of mutual information allows a threshold to be set. This allows TF-target pairs with low MI to be screened out - an MI value will only be returned for a target when it exceeds this value. The threshold can be specified as a mutual information value or as a P-value. (Note - in current implementations of geWorkbench, the p-value calculation is not available for the conditional MI calculation, and we recommend that a MI value of around 0.2 be tried. The unconditional MI is intended for use in the calculation of statistical significance of the final delta MI score and is not currently used.
- Mutual Info - If selected, the user specifies a threshold for the mutual information (MI) estimates in terms of the raw MI score. For example, a value of 0.20 filters out target genes with a MI score of less than 0.20. By default, a MI threshold of 0.1 is set.
- P-value - If selected, the user specifies a threshold for the mutual information estimate in terms of a p-value - an estimate of the significance of the value. This is a value between 0 and 1, with 1 indicating no threshold. By default, the value is 0.01.
- NOTE - p-value is not currently offered for the conditional calculation. The p-value for the unconditional calculation is a rough estimate only. Full p-value calculations will be implemented in a future release of MINDy within geWorkbench.
- Correction - None or Bonferroni - correct for multiple testing if a p-value is specified.
- DPI Tolerance - The Data Processing Inequality (triangle inequality)can be used to remove the effects of indirect interactions, e.g. if TF1->TF2->Target, DPI can be used to remove the indirect action of TF1 on the target. Stated another way, the DPI can be used to remove the weakest interaction of those between any three markers. The DPI tolerance specifies the degree of sampling error to be accepted, as with a finite sample size an exact value MI can not be calculated.
- The DPI tolerance is normally between 0 and 0.15 since values larger than 0.15 yields higher false positives.
- See the Tutorial_-_ARACNE tutorial page and Margolin et al. 2006 for further details on use of DPI.
- DPI Target List - The DPI target list can be used to limit the ARACNE calculation to transcriptional networks. It is used to screen out spurious regulatory interaction signals of genes that are tightly coexpressed but are not in a regulatory relationship to each other, for example genes for two proteins that are in a physical complex and hence always produced in the same amounts. A comma-separated list can be typed in, or it can be loaded from an external file. If used, the DPI Target List should contain all markers that are annotated as transcription factors. Signaling proteins could also be included.
- Details: If the box is checked, the user selects and loads a file which specifies markers (which should be a list of one or more presumptive transcription factors) which will be given preferential treatment during the DPI edge-removal step. Edges originating from markers on this list will not be removed by edges originating from markers not on this list. However, for DPI calculations where all three markers are members of the list, the weakest connecting edge may still be removed.
Services (Grid)
MINDy can be run either locally within geWorkbench, or remotely as a grid job on caGrid. See the Grid Services section for further details on setting up a grid job.
Running a MINDy Analysis
1. Select a microarray set node in the Project Folder.
2. In the analysis pane (lower right of the application), select MINDy Analysis.
3. In the Main tab, populate the Modulators List by selecting a set of markers defined in the Markers component, or load a list from a file.
4. Populate the Target List textbox by selecting the choice "All Markers", or by selecting a set of markers defined in the Markers component, or by loading a list from a file.
5. Populate the Hub Gene textbox to designate the TF gene by (1) typing the marker name (as displayed in the Markers component) or (2) in the Selection Area (lower left of the application) Marker Tab, click on the marker name corresponding to the TF.
6. Parameter values for the unconditional and conditional mutual information calculations can be set in the Advanced Tab. The values will depend on the specifics of the data set being used, in terms of number of arrays and number of markers. A suggested "first try" set of parameters as shown in the above screenshot of the "advanced parameters" tab is:
- sample per condition: 35%
- conditional: MI 0.1 (or even 0)
- unconditional: MI 0.1
- DPI target list: blank
- DPI tolerance: 0.1
7. Click Analyze. If successful, the project window is updated to reflect the MINDy result node. The result node is shown as a child node of the input dataset. Please note that the Dataset History tab captures the analysis parameters.
Viewing MINDy Results
1. Select the MINDy result node in the Project Folder.
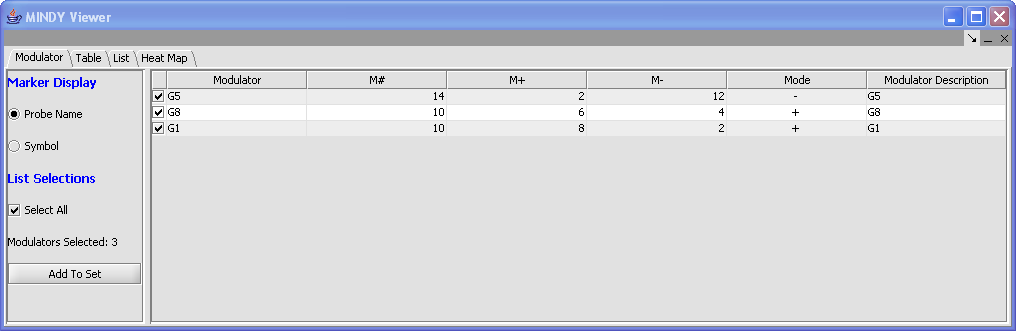
2. In the Modulator Tab, indicate the modulators of interest using the checkboxes or click on Select All to display all modulators in the Table, List,and Heat Map views. The Modulators Selected is updated to reflect the number of modulators selected. Only selected Modulators are displayed on the Table, List and Heat Map views. Additional actions include:
- Marker Display: Indicate marker display preferences for the Modulator column ( probe name or symbol).
- Sort: Click on the column headers or use sort options available in the left pane.
- Add to Set: Adds selected modulators to a Marker Set. You can select one or more Targets and/or Modulators, using the selection checkboxes.
- All Markers: This checkbox determines if all the target genes are displayed or only genes in activated marker groups.
3. Select from the various tabs to view the data in alternate formats. See [#_Navigating_MINDy_Visualization Navigating MINDy] for additional information on these data views.
MINDy includes the following data views: Modulator, Table, List and Heat Map.
Modulator
Modulator: This table-based view contains one row per modulator gene. Only modulators selected in this tab are included the other data views. The value of the Mode column for a modulator M is either “+”, “-“ or null (0) depending on if M+ is larger, smaller or equal to M-.
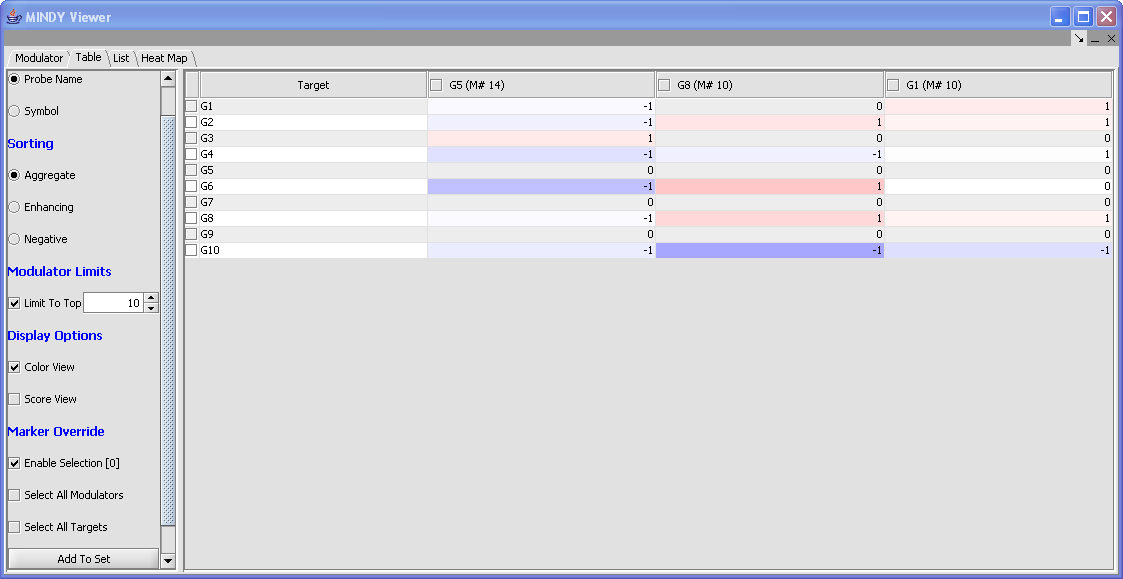
Table
Table: The rows of the table represent target genes and the columns represent modulators. Additional actions include:
- Marker Display: Indicate marker display preferences for the Modulator column ( probe name or symbol).
- Sorting: Displays columns (modulators) from left to right in descending order by; Aggregate ( M#), Enhancing (M+) or Negative (M-).
- Modulator Limits: Activates the checkbox to limit the columns (modulators) display to a defined value. This selection filters the modulator display based upon the current display order.
Display Options:
- Color View: Enables a heat map display of each cell based on the value of the score. 1 is displayed as absolute blue; +1 is displayed as absolute red; 0:1 is mapped uniformly from white to shades of red; -1:0 is mapped uniformly from shades of blue to white.
- Score View: Displays the discretized score values.
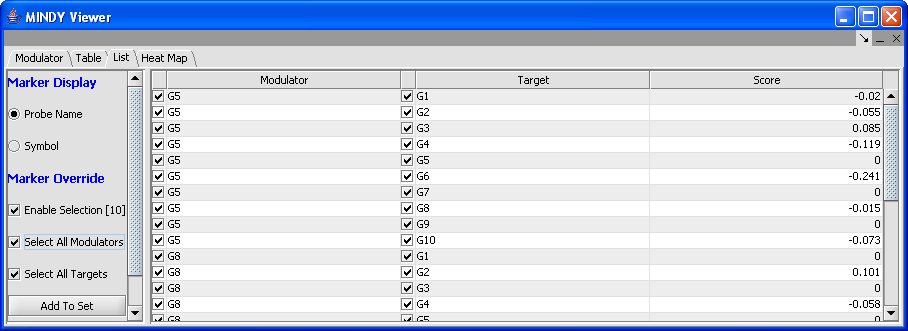
List
List: The table has three columns: Modulator, Target and Score. Additional actions include:
- Marker Display: Indicate marker display preferences for the Modulator column ( probe name or symbol).
- Marker Override: Marker selection preferences. As markers are selected, the number of markers selected is listed next to Enable Selection field. This does not reflect the number of rows.
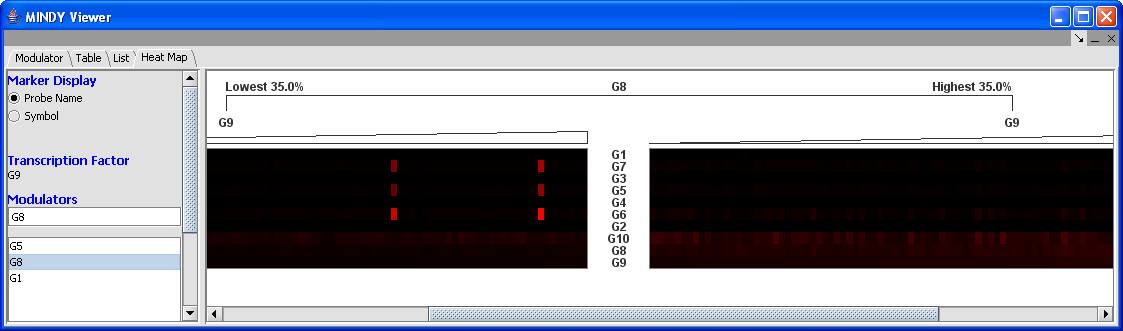
Heat Map
Heat Map: The Heat Map represents the expression values for individual markers (target genes). It contains two color mosaic panels. The rows correspond to target genes and the columns (arrays) are ordered according to the expression of the TF gene (low to high). In the screenshot above, G8 represents the modulator, whose expression values are used to divide the data into two sets. G9 represents the TF whose interaction with various targets is being evaluated. The left panel correspond to the L- arrays where the modulator is least expressed while the columns on the right panel to the L+ arrays where the modulator is most expressed. Additional actions include:
- Marker Display: Indicate marker display preferences for the Modulator column ( probe name or symbol).
- Transcription Factor: Displays the TF entered in the MINDy Analysis parameters.
- Modulator: Select a modulator from the “Selected Modulators” list to update the heat map display.
- Refresh: Resets the heat map display.
- Image Snapshot: Captures the heat map as an image node in the Project Folder.
References
Margolin, A., Wang, K., Lim, W.K., Kustagi, M., Nemenman, I., and Califano, A. Reverse Engineering Cellular Networks. Nature Protocols, 2006 Vol 1(2). ppgs 662-671.'
Wang, K. et al., (in preparation) MINDY: An Algorithm for the Genome-wide Discovery of Modulators of Transcriptional Interactions. See http://arxiv.org/PS_cache/q-bio/pdf/0510/0510030v2.pdf