Array Sets
Contents
Outline
In this tutorial, you will learn
- How geWorkbench allows sets and subsets of microarrays to be created.
- How to mark a subset of arrays as "Active"
- How to classify a subset of arrays, e.g. as "case" vs. "control".
Overview of Marker and Array Sets
The Markers/Arrays component, located at lower left in the geWorkbench graphical interface, allows the user to define and use subsets of arrays and markers for a number of purposes.
As used in geWorkbench, the term "marker" includes genes, probes/probesets, and individual sequences, depending on the type of data loaded. Sets of markers can be returned by various analysis routines. For example the t-test returns a list of markers showing significant differential expression, and after hierarchical clustering, the markers in a subtree of the resulting dendrogram can be saved to a list.
Sets of microarrays can be used to group arrays in a meaningful fashion for statistical analysis. For example, two such phenotypes might be the diseased and normal states of a tissue from which samples have been taken. geWorkbench uses the terms "Case" and "Control" to categorize these, but in biological setting the equivalent would be "Experimental" vs "Control".
This chapter discusses the use of subsets of microarrays. Please see the chapter Data Subsets - Markers for a discussion of the use of Marker sets.
Common Principles of Operation of Marker and Array Subsets
Rather than using all arrays or all markers in a data set for a particular analysis or visualization, the user may wish to restrict those used to only some subset.
Activating Subsets of Markers and Arrays
In the Markers and Arrays components, subsets of markers and arrays can be defined by the user, and also are created as the outcome of some analyses. Beside each such subset in the graphical interface is a checkbox. Marking this box "checked" activates the subset.
- Activating a subset restricts many geWorkbench components to using as input only the markers or arrays that are in such activated subsets.
- Marker Subsets
- If no Marker subset is active, all Markers are used.
- If at least one Marker subset is activated, affected components will only use markers in activated sets.
- Array Subsets
- If no Array subset is active, all Arrays are used.
- If at least one Array subset is activated, affected components will only use arrays in activated sets.
Preparation
In this tutorial we will start with the same data files that were used in Tutorial - Projects and Data Files. Load the ten individual MAS5 data files as shown there in the section "Creating a new project and loading microarray data files".
Assigning arrays to sets
We will place the arrays in the default group, however you can create a new group by pushing the New button on Array/Phenotype Sets at lower left.
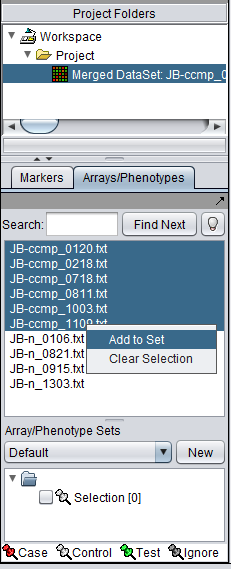
First, we will select and label arrays which contain samples from the congestive cardiomyopathy disease state:
1. In the Arrays/Phenotypes component, select the six arrays beginning with JB-ccmp, which represent the samples from the congestive cardiomyopathy disease state.
2. Right-click, select Add to Set.
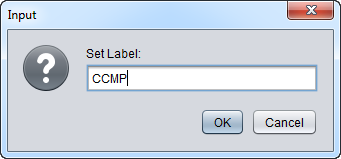
3. Enter "CCMP" in the input box and click OK.
4. Next, similarly label the arrays beginning with JB-n as "Normal" ( repeat steps 2 & 3 ):
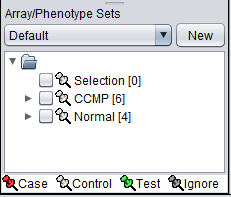
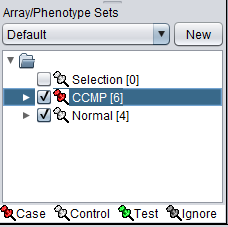
The Array/Phenotype Sets component will now show the two subsets added:
Activating subsets
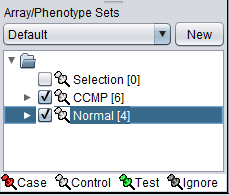
The check boxes next to the subset name can be checked to indicate that a subset of arrays is "Active". Various analysis and visualization components can be set to only use/display activated arrays or markers.
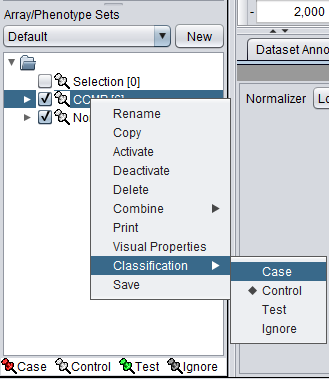
Classifying a subset
For statistical tests such as the t-test, Case and Control sets can be specified.
- Left-click on the thumb-tack icon in front of the phenotype name.
- Select Case to specify the disease arrays as the "Case". The remaining "Normal" arrays are by default labeled control.
A red thumbtack indicates the arrays have been specified as "Case".
Example of working with multiple array sets
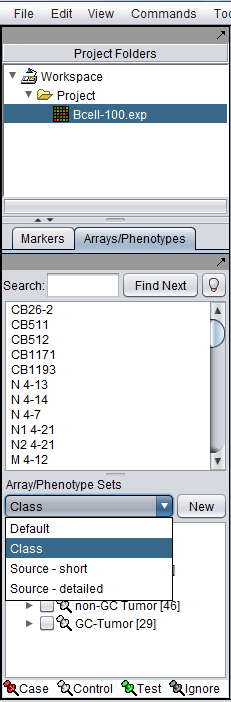
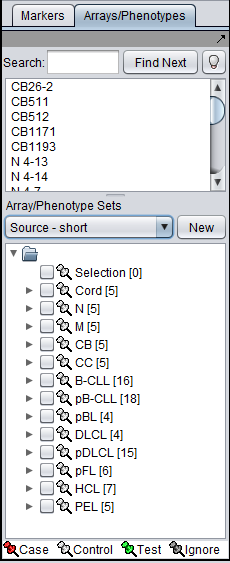
There can be different groupings of the same arrays in the Arrays/Phenotypes and Marker components. Here we show how there are several different set groupings which are predefined in the example data file "BCell-100.exp". After loading this file into geWorkbench as type "Affymetrix File Matrix", the following sets can be seen in the Arrays/Phenotypes group pulldown menu:
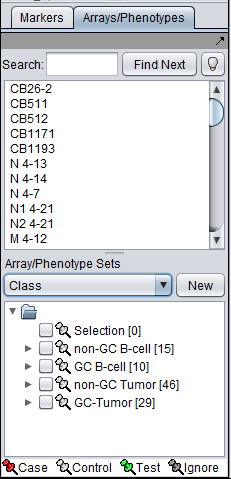
If we choose the set called "Class", the following subsets of arrays are displayed:
If instead we choose the set "Source - short", a different division into subsets of the same arrays is seen: