Gene Ontology Viewer
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
The Gene Ontology Analysis Viewer allows direct browsing of the Gene Ontology, and also the visualization of GO Term analysis results.
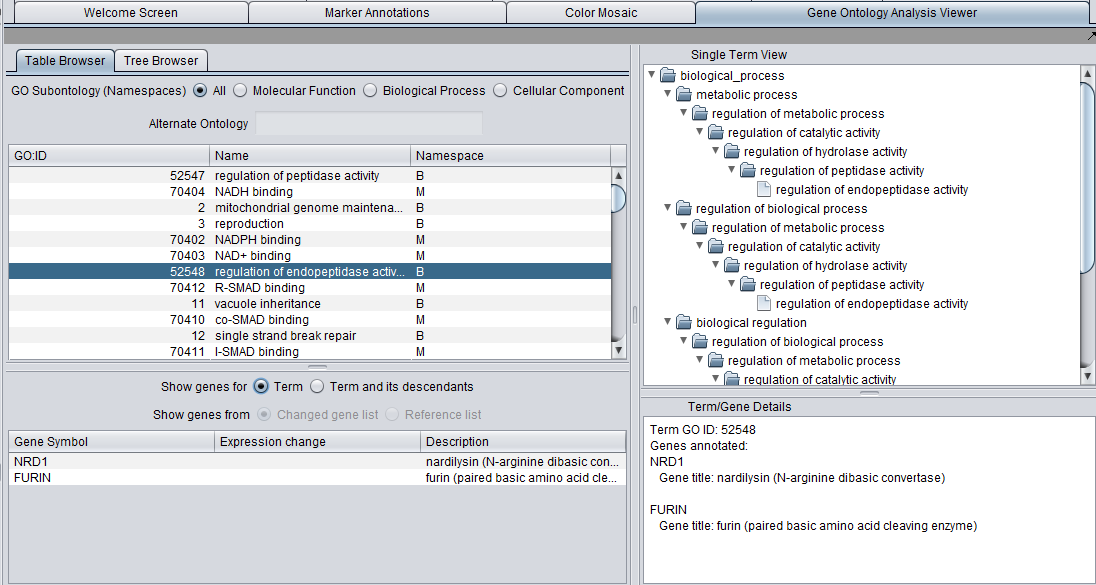
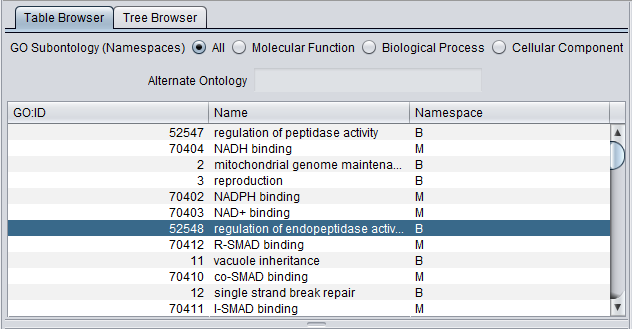
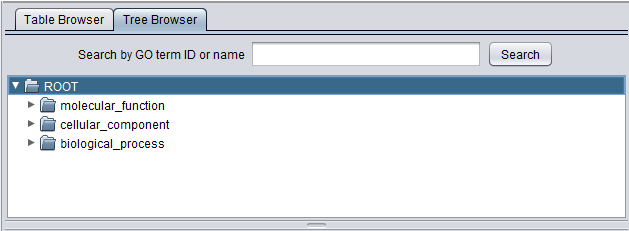
The viewer presents the GO both in tabular form (Table tab) as well as in a tree form (Tree tab).
In addition, three windows provide additional details:
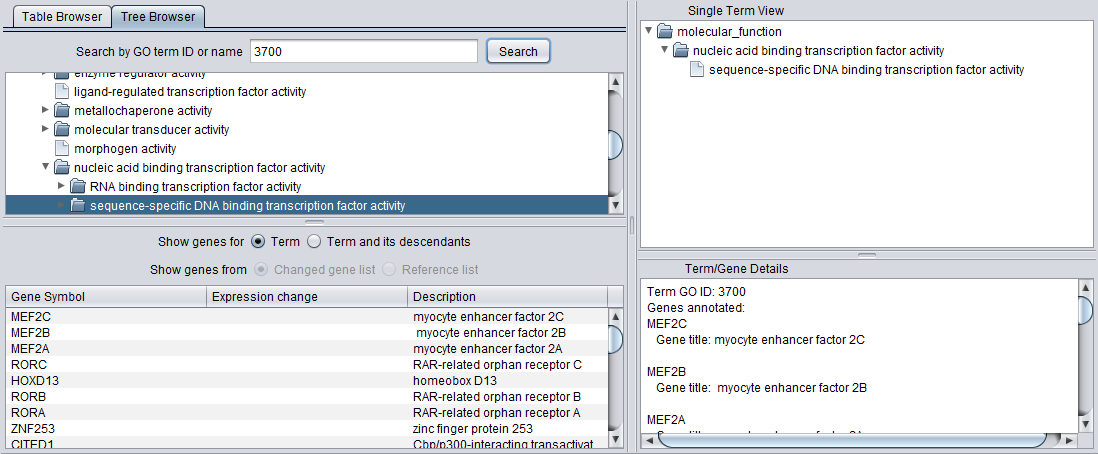
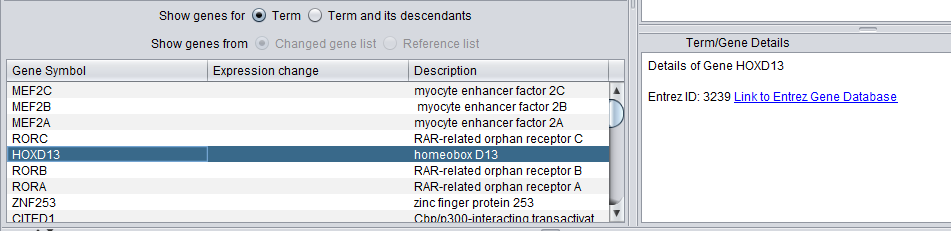
- A list below the selection windows shows all genes annotated with a selected term.
- The "Single Term View" shows the complete tree, including multiple paths through the GO directed acyclic graph, for any selected term.
- The Term/Gene Details windows provides additional details on an term or gene selected in the GO browser or the gene list.
Standalone Viewer Details
Table Browser
Tree Browser
Gene List