CaArray
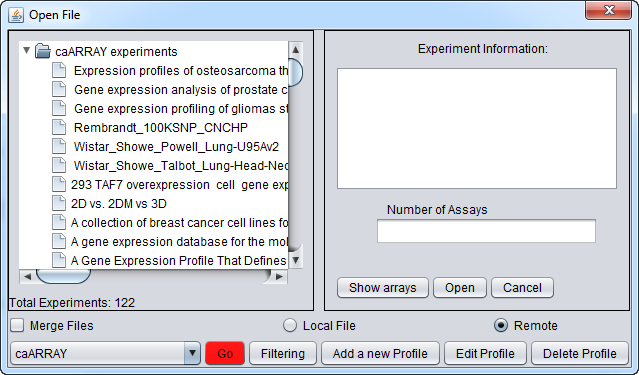
The Remote Open File dialog (caArray)
geWorkbench can retrieve gene expression data from remote instances of the NCICB's caArray database. These may be copies maintained by the NCI itself, or copies maintained locally at your or another institution. You can maintain settings for any number of different caArray installations.
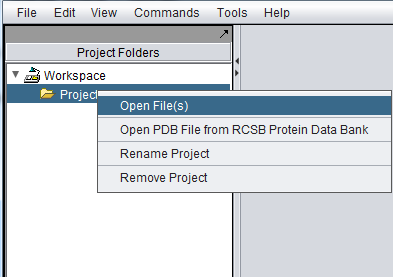
To connect to a remote caArray, right-click on Project in the geWorkbench main window,
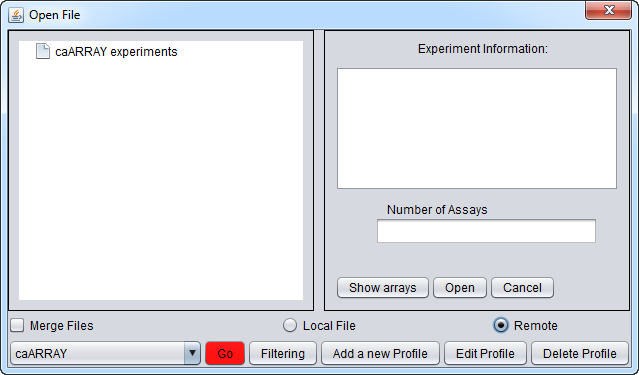
which will bring up the Open File(s) dialog. Click the Remote radio button. The Open File dialog window will be expanded to include remote caArray sources as shown here.
Connection and Query Controls
The buttons at the bottom of the remote file dialog are:
- caArray (Source) menu - Shows a list of caArray instances that have been configured. Entries for the "Production" and "Stage" instances of caArray maintained by NCI are preconfigured.
- Go button - downloads a list of all available experiments from the remote source.
- Filtering - The list of available experiments can be "filtered" to show only those matching particular criteria.
- The available filtering options are:
- Categories: Experiment
- Field Selection: Array Provider, Organism, Principal Investigator
- Values:
- Array Provider: Affymetrix, Agilent, GenePix, Illumina, ImaGene, Niblegen, ScanArray, UCSF Spot.
- Organism: many entries, including human, mouse, fly etc...
- Principal Investigator: PIs of listed public experiments in the particular instance of caArray.
- Add A New Profile button - Opens the Data Source Definition Page used to add a new instance of caArray.
- Edit Profile button - Edit the currently selected profile.
- Delete Profile - Remove the currently selected profile.
Experiment Selection and Download Controls
- caArray Experiments - This pane will contain the list of expriments retrieved from a particular instance of caArray.
- Experiment Information - clicking on any experiment in the list will show its details here.
- Number of Assays - The number of hybridizations for which data is available in this experiment, only available after "Show Arrays" has been pushed.
- Show Arrays - Retrieve the full list of available hybridizations for this the selected experiment.
- Open - Download the hybridizations which have been selected in the list.
- Cancel - Dismiss the Remote dialog.
Loading data from an instance of caArray
Setting up the connection
On the Open File dialog, you can use
- Add a New Profile to set up a new connection to an instance of caArray. Use
- Edit Profile to change the settings of an existing profile.
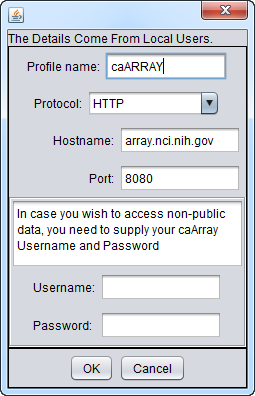
The settings for the production instance of caArray at the NCI are shown here:
- Profile Name - assigns a name to the profile
- Protocol - Method by which to communicate with the remote caArray server.
- HTTP - this is the only protocol currently supported.
- HTTPS - not used.
- RMI - not used.
- Hostname - the URL for the desired caArray server.
- Port - the port on the remote caArray server at which caArray is available.
- Username and Password (optional) - caArray supports retrieval of data from private experiments. These fields allow the user to provide his or her caArray credentials to gain access to any private experiments to which he or she has rights. No username or password is needed for accessing public experiments.
- Note - Once a username and password have been entered and submitted to caArray, you cannot go back to using no username/password, except by restarting geWorkbench. However you can still put in a different username/password combination. This is a property of the caArray server-side code. Thus if you have no valid username/password and enter an incorrect one, you will need to restart geWorkbench before you can query caArray public experiments again (no login required).
Searching and viewing available experiments
If you click on the red Go button next to the caArray data source at the bottom of the dialog, all available caArray experiments at that location will be displayed.
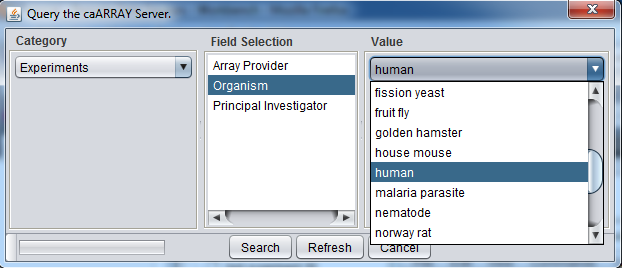
Instead, you can select only particular kinds of experiments by pushing the Filter button. Here we show experiments of type "Human" being selected.
And here are the resulting entries in the database:
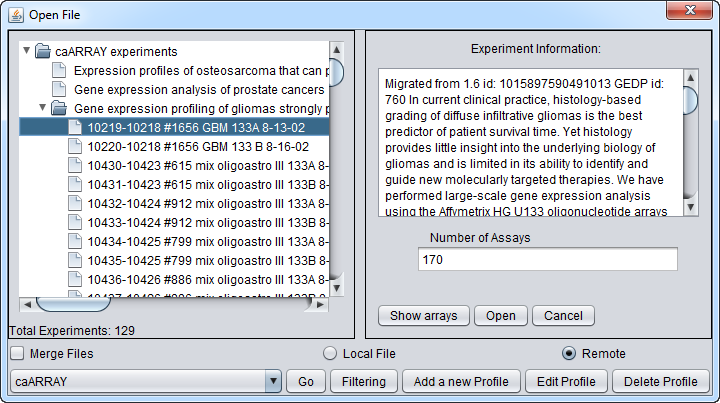
Viewing the list of arrays available in a given experiment
Select an experiment and push the Show Arrays button to see the individual array datasets available for download for that experiment.
(As of geWorkbench v2.2.2, the list of arrays will be displayed sorted alphabetically.)
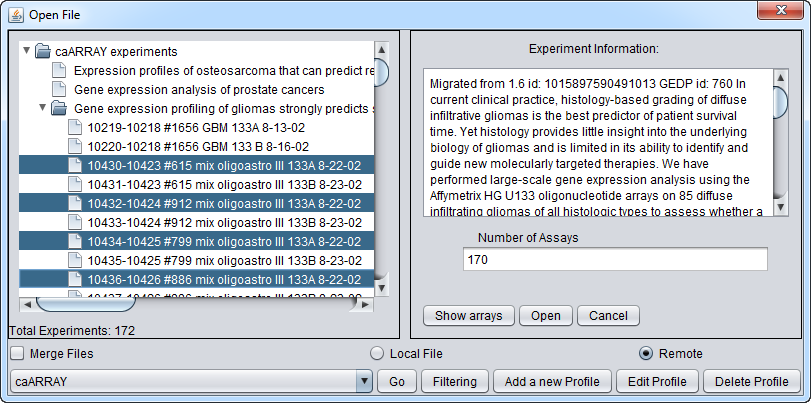
Downloading select array datasets
Now we will select four of the arrays of type HG-U133A and push the Open button to begin the download. Dont' forget to click the Merge button first if desired to merge the data into a single dataset.
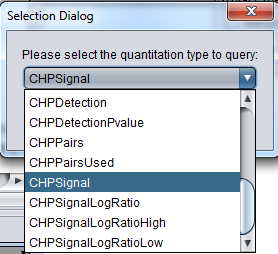
You will be prompted to select the quantitation type from those available for the experiment. Although all available quantitation types available for the dataset are listed, usually only the signal channel should be used. Here we select CHPSignal (Affymetrix derived data CHP file):
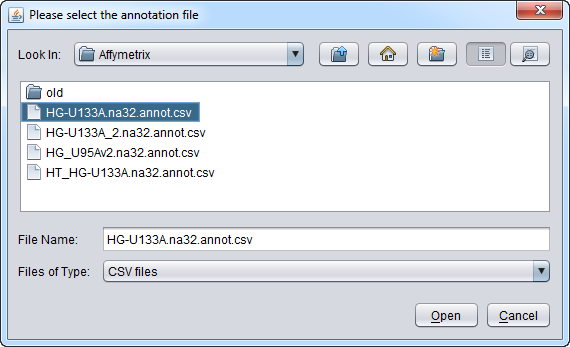
You will also be prompted to load an annotation file, if desired. Here we choose the Affymetrix HG-U133A annotation file.
A progress bar will track the download process:
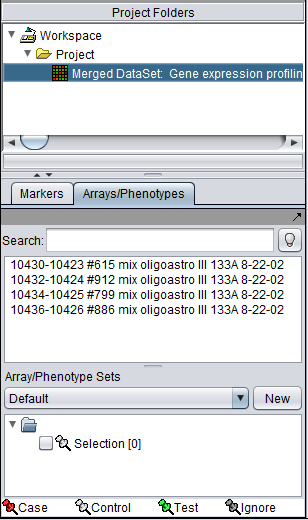
The resulting data set will appear in the Project Folders component, and the individual arrays are listed in the Arrays component: