Difference between revisions of "ANOVA"
(→Overview) |
|||
| Line 7: | Line 7: | ||
| − | Image:T_ANOVA_array_groups.png | + | [[Image:T_ANOVA_array_groups.png]] |
| − | Image:T_ANOVA_ColorMosaic.png | + | [[Image:T_ANOVA_ColorMosaic.png]] |
| − | Image:T_ANOVA_setup.png | + | [[Image:T_ANOVA_setup.png]] |
| − | Image:T_ANOVA_significant_genes_set.png | + | [[Image:T_ANOVA_significant_genes_set.png]] |
Revision as of 12:41, 28 July 2009
Overview
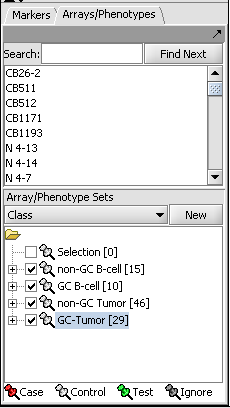
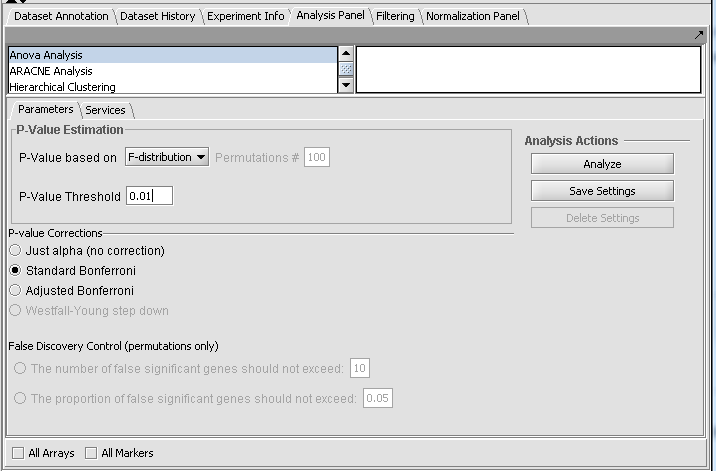
The geWorkbench ANOVA component utilizes a one-way analysis of variance calculation from TIGR's MEV package. At least three groups of arrays must be specified by defining and activating them in the Arrays/Phenotypes component. For each chosen marker the routine determines if, at the specified level of significance, any difference exists in expression values between any of the groups (the null hypothesis is that there is no difference between the groups). Several basic methods of multiple testing correction are offered. The analysis does not indicate between which groups the difference is found, only that one exists.
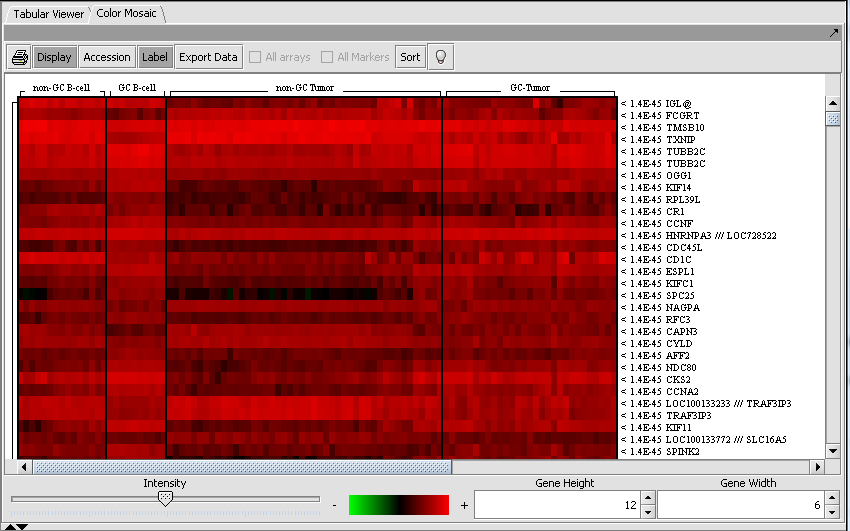
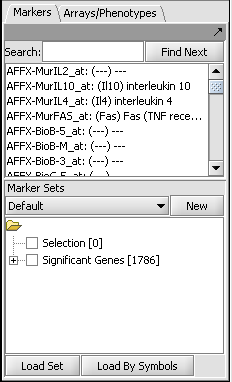
Those markers for which a significant difference is found are placed into a new set in the Markers component called "Significant markers". The results are also display as a heat map in the Color Mosaic component.