Difference between revisions of "ANOVA"
(→Setting up ANOVA Parameters) |
(→Setting up ANOVA Parameters) |
||
| Line 75: | Line 75: | ||
[[Image:T_ANOVA_array_groups.png]] | [[Image:T_ANOVA_array_groups.png]] | ||
| − | ==Setting up ANOVA | + | ==Setting up the Parameters and running ANOVA== |
For this example we wil apply a relatively stringent multiple testing correction. | For this example we wil apply a relatively stringent multiple testing correction. | ||
| − | # Leave the P-value method set to F-distribution. | + | # Leave the P-value method set to '''F-distribution'''. |
| − | # Set the | + | # Set the '''P-Value Threshold''' (alpha) to 0.01. |
| − | # For the P-value correction choose Standard Bonferroni. | + | # For the P-value correction choose '''Standard Bonferroni'''. |
| + | # Push the '''Analyze''' button. | ||
Revision as of 14:55, 28 July 2009
Contents
Overview
The geWorkbench ANOVA component utilizes a one-way analysis of variance calculation from TIGR's MEV package. At least three groups of arrays must be specified by defining and activating them in the Arrays/Phenotypes component. For each chosen marker the routine determines if, at the specified level of significance, any difference exists in expression values between any of the groups (the null hypothesis is that there is no difference between the groups). Several basic methods of multiple testing correction are offered. The analysis does not indicate between which groups the difference is found, only that one exists.
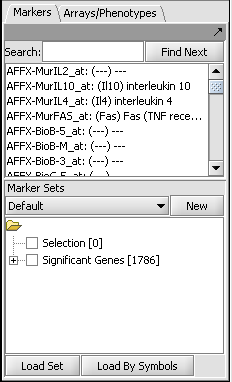
Those markers for which a significant difference is found are placed into a new set in the Markers component called "Significant Genes". The results are also display as a heat map in the Color Mosaic component.
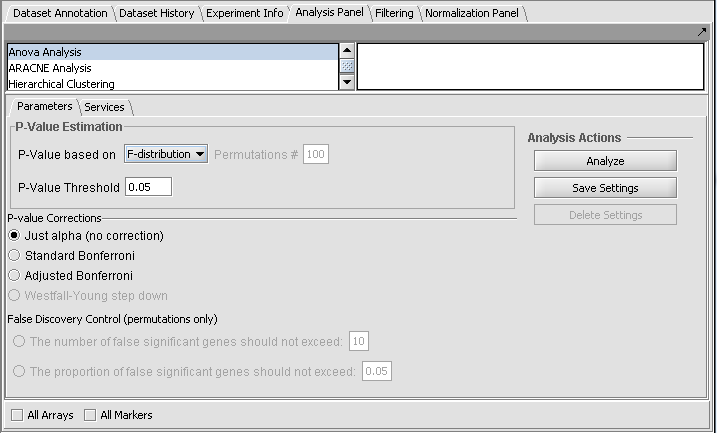
ANOVA Parameters and Settings
P-Value Estimation
F-Distribution
The F-distribution arises from the ratio of the variances of two normally distributed statistics (chi-squared distributions).
Permutation
This selection requires the number of permutations. The default value is 100.
Multiple testing corrections
The p-value can be used directly as a cutoff for determining significant genes, or it can be corrected.
Just Alpha
No correction is performed.
Standard Bonferroni
The cutoff value (alpha) is divided by the number of tests (genes) before being compared with the calculated p-values.
Adjusted Bonferroni
Implements the Holm-Bonferroni correction. For each successive P-value in an ordered list of P-values, the divisor for alpha is decremented by one and then the result compared with the P-value. The effect is to slightly reduce the stringency of the Bonferroni correction.
Westfall-Young Step Down
(This correction is only available when the permutation method is chosen for calculating p-values).
False Discovery Control
(This correction is only available when the permutation method is chosen for calculating p-values).
The user must select the false discoveries in terms of either
- An upper limit on the number of false positives (markers falsely called as showing a significant difference), or
- An upper limit on the proportion of false positives.
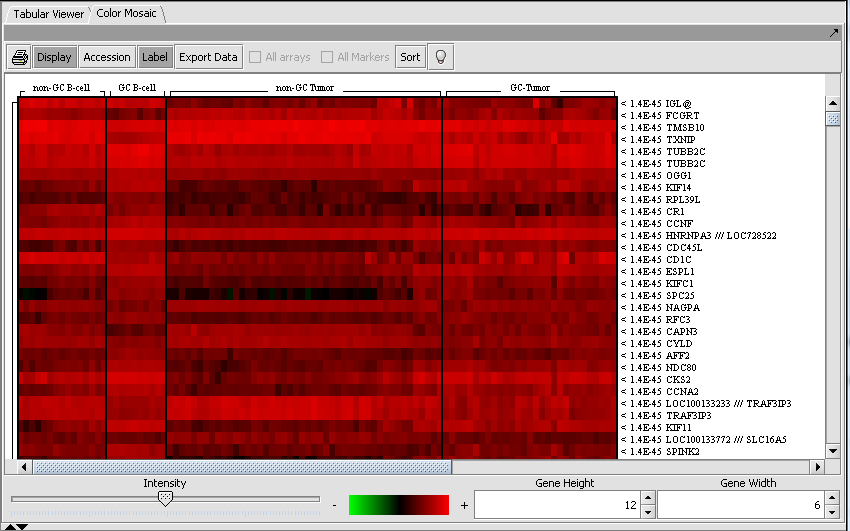
Viewing ANOVA Results
When an ANOVA result node is selected (highlighted) in the project panel, the results will be displayed in both a tabular form and in the form of a heatmap in the Color Mosaic component.
Color Mosaic
In this view, a color spectrum is used to indicate the relative magnitudes of the measurements. The arrays (columns) are grouped by input group membership, i.e. group 1, group 2 etc. Each row corresponds to a marker, and markers display is ordered by p-value ascending order (from most significant to least significant). This visualization is described in detail in the Color Mosaic Help and Tutorials.
Tabular View
This Visual Area component displays a read-only spreadsheet view of the significant genes sorted by p-value in ascending order (from most significant to least significant).
Basic Navigation
- Resize columns by using the mouse to drag column headings.
- Reorder columns in the details pane by using a mouse to drag a column heading to the left or right of its original position. As you drag a column, highlighting between the column headings indicates the new position of the column.
- To sort by a specific column, double click on the column header to sort ascending and then click again to sort in descending order
- Display Preference: Display Preference button to modify the visible columns.
- Export: Click on Export in the lower left of the visualization to export this table in .csv format. The export file will contain only the columns displayed.
Example of running ANOVA
Loading the example data
This example uses the Bcell-100.zip dataset available and further described on the Download page. Briefly, this dataset is composed of 100 Affymetrix HG-U95Av2 arrays on which various B-cell lines, both normal and cancerous, were analyzed. Thus it explores a potentially wide variety of expression phenotypes. In order to see the gene names associated with the probesets, one must also download the correct annotation file for this array from the Affymetrix Netaffx website.
- Obtain the Bcell-100.exp dataset. If downloaded as a zip file, unzip it to a convenient directory.
- (Optional) Obtain the annotation file for the HG-U95Av2 array type from Affymetrix. The name will be similar to "HG_U95Av2.na28.annot.csv", where na28 is the version number.
- Load the Bcell-100.exp dataset into geWorkbench as type "Affymetrix File Matrix". (See Loading from File).
- When prompted, and if desired, load the annotation file.
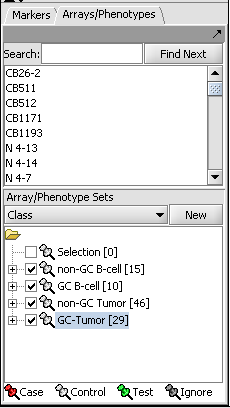
Choosing array groups
The Bcell-100 dataset comes with predefined sets of arrays. In the Arrays/Phenotypes component (at lower left in the geWorkbench GUI), choose the group in the pulldown menu called "Class". Check the box beside each of the four sets of arrays to activate them as shown in the figure below.
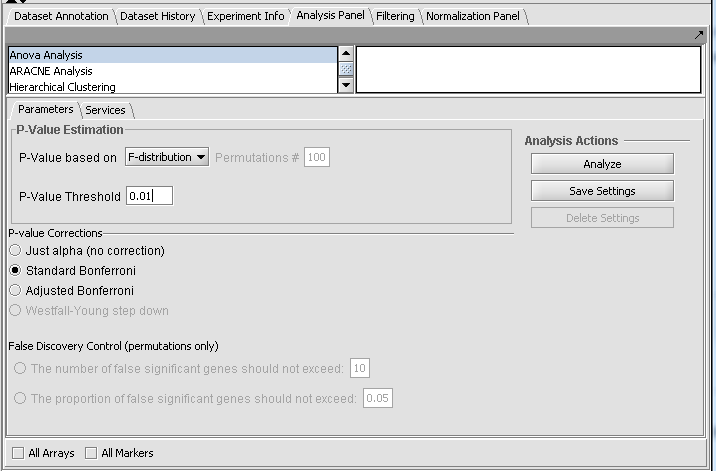
Setting up the Parameters and running ANOVA
For this example we wil apply a relatively stringent multiple testing correction.
- Leave the P-value method set to F-distribution.
- Set the P-Value Threshold (alpha) to 0.01.
- For the P-value correction choose Standard Bonferroni.
- Push the Analyze button.
ANOVA result display