Difference between revisions of "BLAST"
(→Example) |
(→Technical Notes) |
||
| (224 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{TutorialsTopNav}} | {{TutorialsTopNav}} | ||
| − | = | + | =Overview= |
| − | + | "The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families" (quoted from the [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome NCBI BLAST homepage]). | |
| − | + | geWorkbench submits BLAST jobs to the NCBI server. NCBI-supported sequence databases and search algorithms can be selected in the user interface. Since release 2.1.0, geWorkbench supports almost all BLAST setting options available through the NCBI web interface. | |
| − | + | Please note that although, in geWorkbench, we have adopted the default settings for each BLAST algorithm as seen on the NCBI website, those settings are subject to change at any time by NCBI. Before submitting a BLAST job from geWorkbench, the user should verify that the parameter settings are appropriate for their query. | |
| − | |||
| + | The BLAST analysis is available when a protein or DNA sequence is loaded and selected in the [[Workspace|Workspace]]. The BLAST Analysis and BLAST Results Viewer must be loaded in the [[Component Configuration Manager]]. | ||
| − | |||
| − | |||
| − | |||
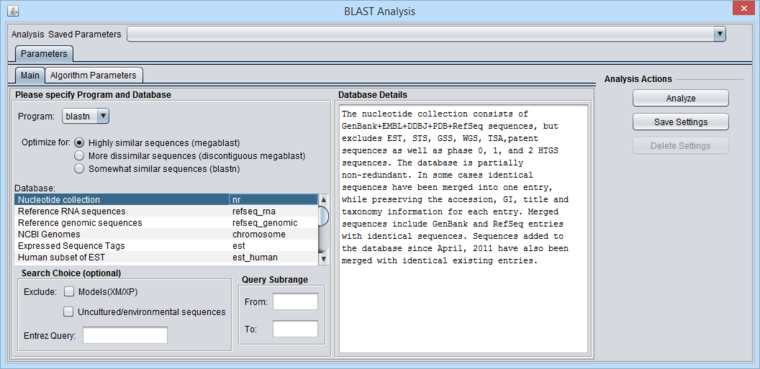
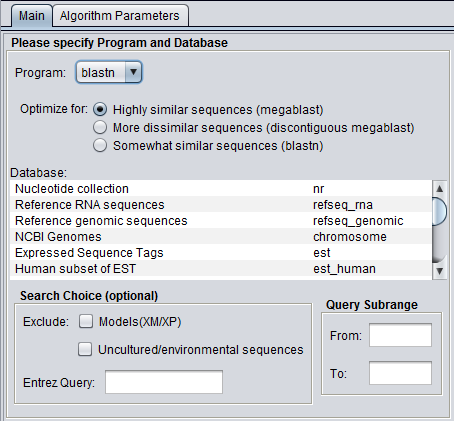
| − | + | [[Image:BLAST_Parameters_Main-blastn-full.png|{{ImageMaxWidth}}]] | |
| − | ===Query | + | '''Figure legend: BLAST Main Parameters'''. A nucleotide sequence has been loaded into the [[Workspace|Workspace]]. |
| − | BLAST accepts nucleotide or amino-acid query sequences in the FASTA format. A query file can contain one or multiple sequences. The file can be loaded from disk using the '''File Open''' command, or may have been placed into the | + | |
| + | =NCBI Documentation= | ||
| + | geWorkbench serves as a interface to the NCBI BLAST server and implements the same options as the NCBI BLAST website. Detailed information about each option can be found on NCBI webpages, including: | ||
| + | * [http://blast.ncbi.nlm.nih.gov/Blast.cgi NCBI BLAST Home Page] | ||
| + | * [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs BLAST Docs - Main help page] | ||
| + | ** [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=ProgSelectionGuide Blast Program Selection Guide] | ||
| + | |||
| + | Older help pages... | ||
| + | * [http://www.ncbi.nlm.nih.gov/blast/html/blastcgihelp.html NCBI BLAST Help page 1] and | ||
| + | * [http://www.ncbi.nlm.nih.gov/BLAST/blastcgihelp.shtml NCBI BLAST help page 2 ]. | ||
| + | |||
| + | |||
| + | |||
| + | =BLAST job setup= | ||
| + | |||
| + | ==Prerequisites== | ||
| + | * The BLAST Analysis and Viewer components must be loaded in the [[Component_Configuration_Manager |Component Configuration Manager]]. | ||
| + | * A protein or nucleotide sequence must be loaded in the [[Workspace|Workspace]]. | ||
| + | |||
| + | ==Query sequences== | ||
| + | BLAST accepts nucleotide or amino-acid query sequences in the FASTA format. A query file can contain one or multiple sequences. The file can be loaded from disk using the '''File Open''' command, or may have been placed into the [[Workspace|Workspace]] by another component such as the Sequence Retriever, or add from the result of a previous BLAST run. | ||
| + | |||
| + | ==Invoking BLAST== | ||
| + | |||
| + | BLAST is a normal geWorkbench analysis component and can be invoked either by right-clicking on a sequence node in the [[Workspace|Workspace]], or through the Commands entry in the Menu Bar at the top of the geWorkbench GUI. | ||
| + | |||
| + | |||
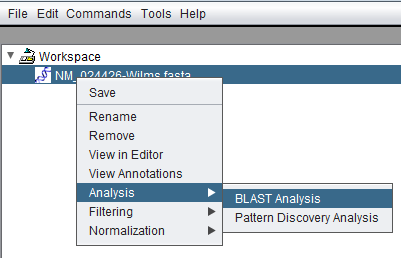
| + | Invoking BLAST via right-clicking on a sequence node in the [[Workspace|Workspace]]: | ||
| + | |||
| + | |||
| + | [[Image:BLAST_Analysis_invocation.png]] | ||
| + | |||
| + | |||
| + | |||
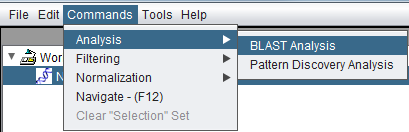
| + | Invoking BLAST via the Commands menu. The desired sequence node must be selected first in the [[Workspace|Workspace]]: | ||
| + | |||
| + | |||
| + | [[Image:BLAST_Analysis_Command_invocation.png]] | ||
| + | |||
| + | ==Parameters - Main== | ||
| + | |||
| + | |||
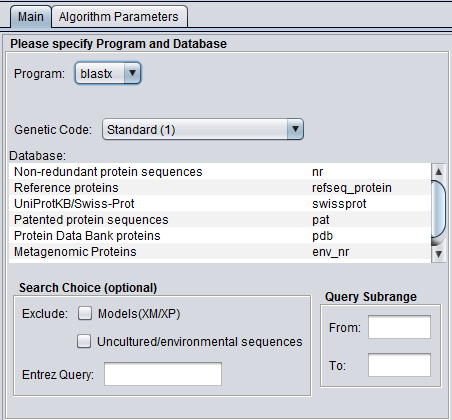
| + | In addition to the parameters shown in the previous image, BLASTX and TBLASTX add the Genetic Code option: | ||
| + | |||
| + | [[Image:BLAST_Parameters_Main-blastx.png]] | ||
| + | |||
| + | ===Algorithms=== | ||
| + | |||
| + | The user must make sure that the algorithm chosen matches the type of query sequence (protein or nucleotide) that has been loaded. Some of the algorithms translate a nucleotide query, a nucleotide database, or both into amino acid sequence before executing the query. Searching in the amino-acid space is more sensitive for certain types of query, as it ignores synonymous, non-functional changes in nucleotide sequence. | ||
| + | |||
| + | ====For protein query sequences:==== | ||
| + | |||
| + | * '''blastp''' - Compares an amino acid query sequence against a protein sequence database. | ||
| + | * '''tblastn''' - Compares a amino acid query sequence against a nucleotide database translated in all reading frames. | ||
| + | |||
| + | ====For nucleotide query sequences:==== | ||
| + | |||
| + | * '''blastn''' - three algorithms are available under the blastn choice, in order of decreasing similarity of the query to the target sequences, Each compares a nucleotide query sequence against a nucleotide sequence database: | ||
| + | ** '''megablast''' - optimize for highly similar sequences. | ||
| + | ** '''discontinuous megablast''' - optimize for more dissimilar sequences | ||
| + | ** '''blastn''' - optimize for somewhat similar sequences. | ||
| + | * '''blastx''' - Compares a nucleotide query sequence translated in all reading frames against a protein sequence database. | ||
| + | * '''tblastx''' - Compares the 6-frame translations of a nucleotide query sequence against the 6-frame translations of a nucleotide sequence database. | ||
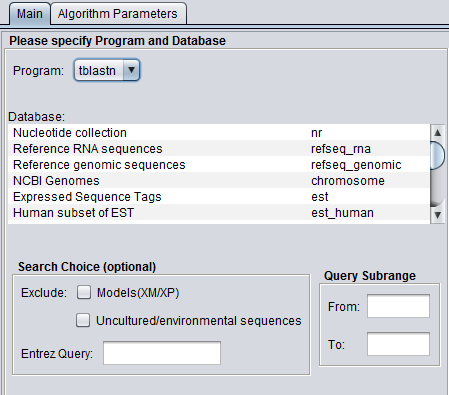
===Databases=== | ===Databases=== | ||
| − | + | Standard protein and nucleic acid databases maintained at NCBI are supported. The appropriate databases for the search algorithm chosen will be displayed. A window to the right, "Database Details", displays a summary of the database currently selected in the list. | |
| + | |||
| + | ====For nucleic acids:==== | ||
| + | * '''Nucleotide Collection (nr/nt)''' | ||
| + | * '''Reference RNA seqeunces (refseq_rna)''' | ||
| + | * '''Reference genomic sequneces (refseq_genomic)''' | ||
| + | * '''NCBI Genomes (chromosome)''' | ||
| + | * '''Expressed Sequence Tags (est)''' | ||
| + | * '''Human subset of EST (est_human)''' | ||
| + | * '''Mouse subset of EST (est_mouse)''' | ||
| + | * '''Non-human, non-mouse ESTs (est_others)''' | ||
| + | * '''Genomic survey sequences (gss)''' | ||
| + | * '''High throughput genomic sequences (htgs)''' | ||
| + | * '''Patent sequences (pat)''' | ||
| + | * '''Protein Data Bank (pdb)''' | ||
| + | * '''Human ALU repeat elements (alu)''' | ||
| + | * '''Sequence tagged sites (dbsts)''' | ||
| + | * '''Whole genome shotgun contigs (wgs)''' | ||
| + | * '''Metagenomic Samples (env_nt)''' | ||
| + | * '''Transcriptome Shotgun Assembly (tsa_nt)''' | ||
| + | |||
| + | ====For proteins:==== | ||
| + | * '''Non-redundant protein sequences (nr)''' | ||
| + | * '''Reference proteins (refseq_protein)''' | ||
| + | * '''UniProtKB/Swiss-prot (swissprot)''' | ||
| + | * '''Patented protein sequences (pat)''' | ||
| + | * '''Protein Data Bank sequences (pdb)''' | ||
| + | * '''Metagenomic proteins (env_nr)''' | ||
| + | * '''Transcriptome Shotgun Assembly (tsa_nr)''' | ||
| + | |||
| + | ===Search Choice=== | ||
| + | These settings provide the ability to restrict the search in certain ways | ||
| + | |||
| + | * '''Exclude''': exclude certain specialized database entries from the search. | ||
| + | ** '''Models''' (XM/XP) | ||
| + | ** '''Uncultured/environmental sequences''' | ||
| + | |||
| + | * '''Entrez Query''' - an Entrez query can be entered directly to restrict a search e.g. to a particular species. Please see [http://www.ncbi.nlm.nih.gov/BLAST/blastcgihelp.shtml NCBI BLAST help]. An example from that page is "Mus musculus[organism] AND biomol_mrna[properties]". This limits the search to mouse mRNA entries in the database. | ||
| − | === | + | ===Genetic Code=== |
| − | + | Genetic code to be used in blastx (and tblastx) translation of the query. | |
| + | |||
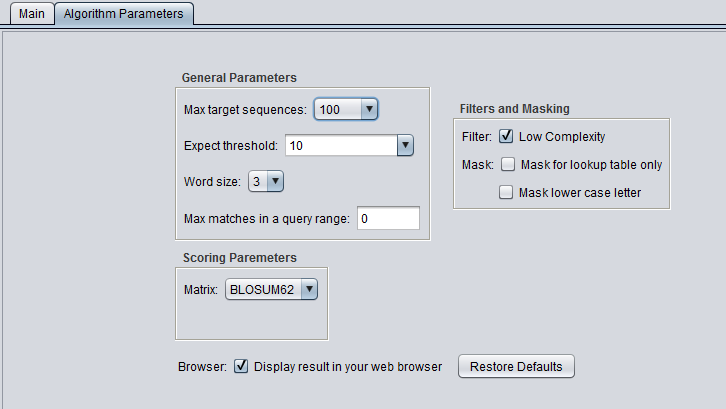
| + | ==Algorithm Parameters== | ||
| + | ===General Parameters=== | ||
| + | * '''Max target sequences''' - maximum number of hits to return. | ||
| + | * '''Automatically adjust parameters for short input sequences'''. | ||
| + | * '''Expect threshold''' - Expected number of chance matches in a random model. | ||
| + | * '''Word size''' - The length of the seed that initiates an alignment. | ||
| + | * '''Max matches in a query range''' - Limit the number of matches to a query range. This option is useful if many strong matches to one part of a query may prevent BLAST from presenting weaker matches to another part of the query. | ||
| + | ** Note - NCBI reports that this feature may not work if there are a large number of full-length sequence matches in the chosen database. | ||
| + | ** The function of this feature is described in [http://www.ncbi.nlm.nih.gov/pubmed/10890403 Berman et al., 2000]. | ||
| + | |||
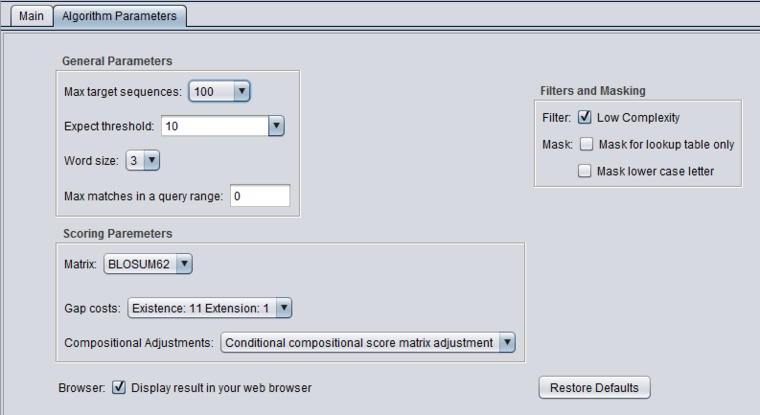
| + | ===Scoring Parameters=== | ||
| + | * '''Match/mismatch scores''' (blastn, megablast, discontinuous megablast) - scores to use for a match or mismatch. | ||
| + | * '''Matrix''' - Various scoring matrices (BLOSUM, PAM) are available for protein and translated queries. | ||
| + | * '''Gap Costs''' - The pull-down menu shows the available choices of gap costs for the current scoring matrix. | ||
| + | * '''Compositional adjustments''' - "...takes into account the amino acid composition of the individual database sequences involved in reported alignments. This improves E-value accuracy, thereby reducing the number of false positive results." | ||
| + | |||
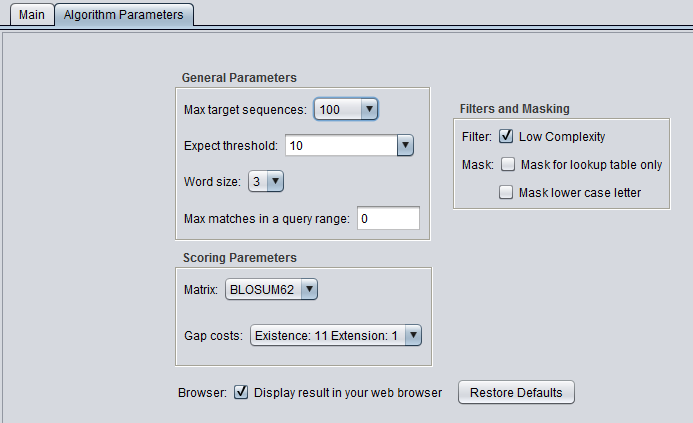
| + | ===Filters and Masking=== | ||
| + | * '''Low Complexity''' - filter out low compositional complexity sequence. | ||
| + | * '''Species-specific repeats filter''' - masks species-specific repeats (e.g. human LINE's and SINE's). This option can speed searches involving long query sequences or databases containing sequences with many repeats. | ||
| + | * '''Mask for lookup table only''' - masks low-complexity sequence only while constructing the lookup table used by the initial hit-find phase of BLAST. The second phase, hit extension, is not not affected and hits can be extended through low-complexity sequence. NCBI notes that this option is experimental and subject to change. | ||
| + | * '''Mask lower case letter''' - filter out sequence which is in lower case in the FASTA query sequence. | ||
| + | |||
| + | ===Discontiguous Word Options=== | ||
| + | |||
| + | Please see the [http://www.ncbi.nlm.nih.gov/blast/discontiguous.shtml NCBI page on discontiguous megablast] for a detailed explanation of these options. | ||
| + | |||
| + | * '''Template Length''' | ||
| + | * '''Template Type''' | ||
| + | |||
| + | ===Other=== | ||
| + | * '''Display result in your web browser''' - geWorkbench will display the HTML page returned by NCBI BLAST in your web browser as well is within its own display. | ||
| + | * '''Restore defaults''' - restore all settings for the currently selected algorithm to their default values. | ||
| + | |||
| + | |||
| + | ===Algorithm Parameter Setting Defaults=== | ||
| + | The default settings for each query type were taken from the NCBI BLAST website. | ||
| + | |||
| + | Please note that options and default settings on the NCBI BLAST website are subject to change at any time. | ||
| + | |||
| + | The user should verify all settings are appropriate for his or her particular BLAST query. | ||
| + | |||
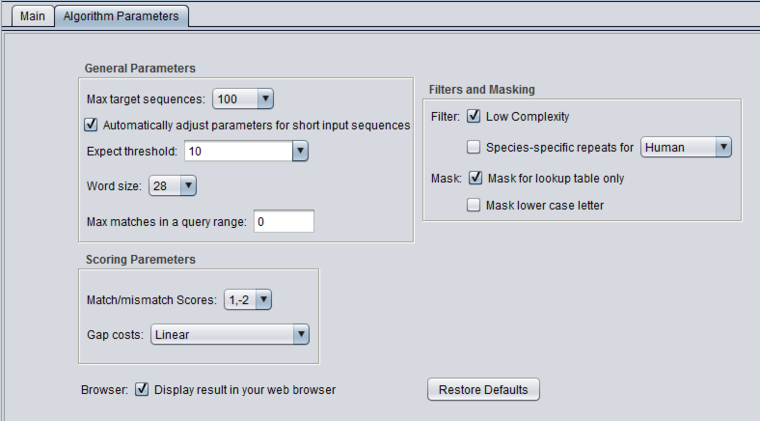
| + | ====blastn - megablast==== | ||
| + | |||
| + | [[Image:BLAST_Parameters_Main-blastn.png]] | ||
| + | |||
| + | |||
| + | [[Image:BLAST_Parameters_blastn-megablast.png|{{ImageMaxWidth}}]] | ||
| + | |||
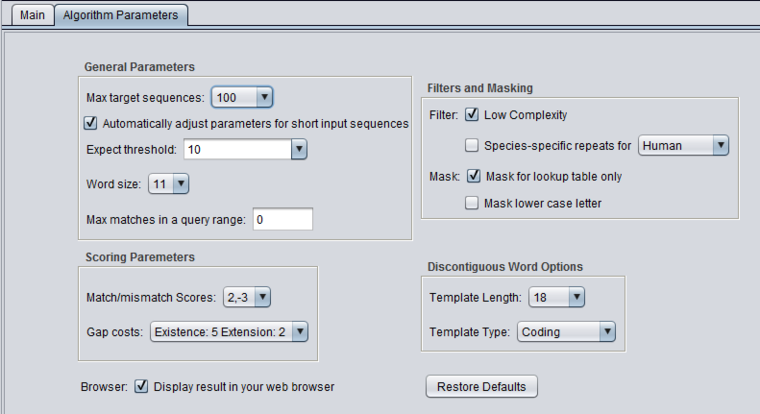
| + | ====blastn - discontiguous megablast==== | ||
| + | |||
| + | [[Image:BLAST_Parameters_blastn-discontinuous-megablast.png|{{ImageMaxWidth}}]] | ||
| + | |||
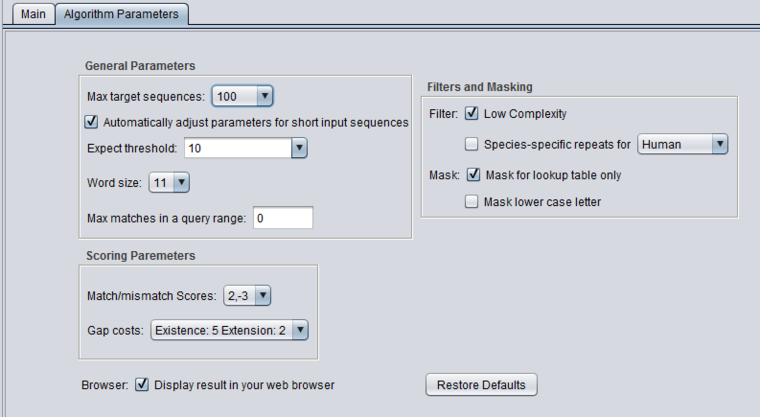
| + | ====blastn - blastn==== | ||
| + | |||
| + | [[Image:BLAST_Parameters_blastn.png|{{ImageMaxWidth}}]] | ||
| + | |||
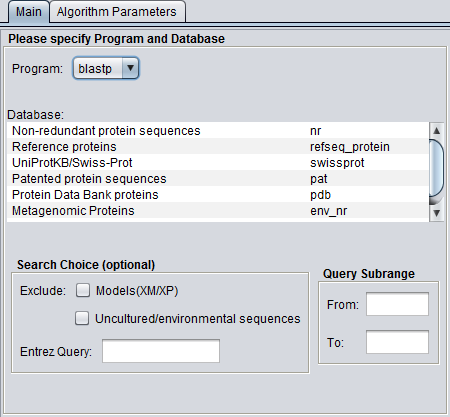
| + | ====blastp==== | ||
| + | |||
| + | [[Image:BLAST_Parameters_Main-blastp.png]] | ||
| + | |||
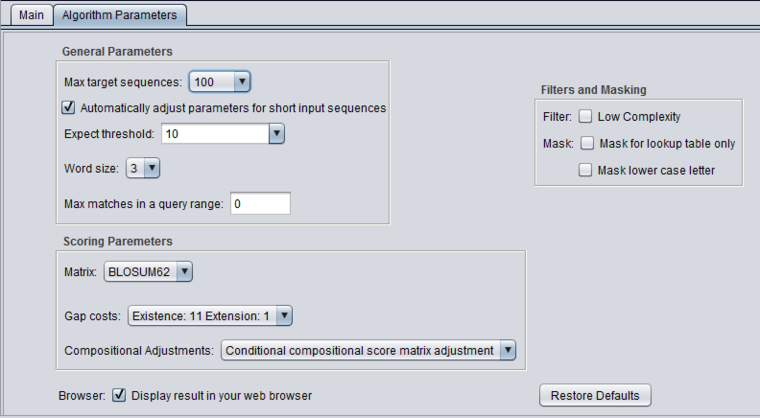
| + | |||
| + | |||
| + | [[Image:BLAST_Parameters_blastp.png|{{ImageMaxWidth}}]] | ||
| + | |||
| + | ====blastx==== | ||
| + | |||
| + | [[Image:BLAST_Parameters_Main-blastx.png]] | ||
| + | |||
| + | |||
| + | [[Image:BLAST_Parameters_blastx.png]] | ||
| + | |||
| + | |||
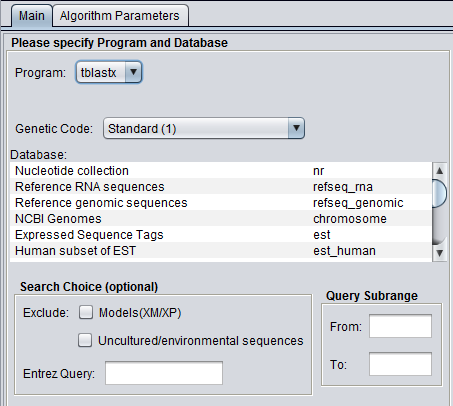
| + | ====tblastx==== | ||
| + | |||
| + | [[Image:BLAST_Parameters_Main-tblastx.png]] | ||
| − | |||
| − | |||
| − | + | [[Image:BLAST_Parameters_tblastx.png]] | |
| − | + | ====tblastn==== | |
| − | + | [[Image:BLAST_Parameters_Main-tblastn.png]] | |
| − | |||
| − | + | [[Image:BLAST_Parameters_tblastn.png|{{ImageMaxWidth}}]] | |
| + | ==Analyze== | ||
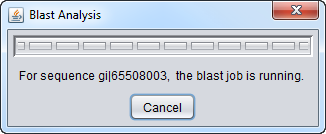
| − | + | The BLAST analysis is launched by pushing the '''Analyze''' button. A dialog with a progress bar will appear. The analysis can be canceled by pushing the '''Cancel''' button on this dialog. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | [[Image:BLAST_Analysis_Progress_Dialog.png]] | |
| − | + | =BLAST Results Viewer= | |
| − | + | When the BLAST search results are returned they are placed in a new node in the [[Workspace|Workspace]] as a child of the query sequence used. Mousing over the result set will show how many sequences are in it. | |
| − | |||
| − | + | [[Image:Blast_result_projects_folder.png]] | |
| − | + | Each different hit is listed on a line in the results table, shown below. Note that a query sequence can hit a database target sequence in more than one place, resulting in multiple alignments displayed per target hit. The results viewer also shows statisics for each hit, including the E-value, start position and length of the hit, and the percent identity. | |
| − | + | If the "Display result in your web browser" option was chosen, then the browser will open with the HTML formated results. | |
| − | + | In the pane at left in the picture below, the name of the input query sequence is shown, e.g. the gi number of a Genbank sequence. If there had been more than one query sequence, then this pane would show a list of query sequence names, allowing you to select the results to be viewed. | |
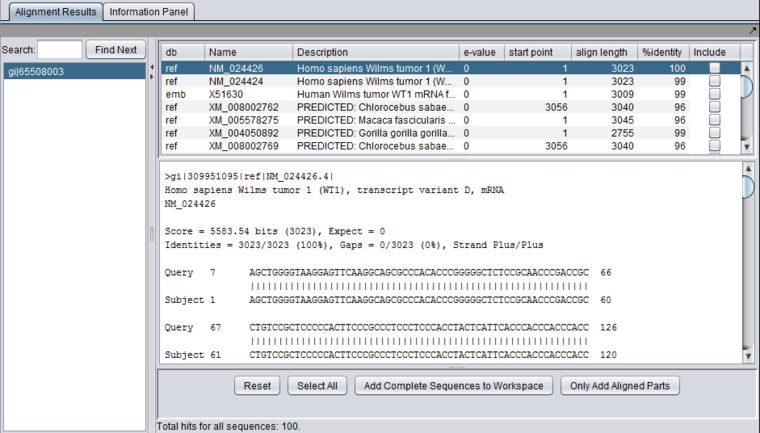
| + | [[Image:Blast_Wilms_NM_result.png|{{ImageMaxWidth}}]] | ||
| + | ==Controls== | ||
| + | ===Within the list of returned hits=== | ||
| + | * '''Include''' check boxes - when checked, selects these sequences for import into the [[Workspace|Workspace]]. | ||
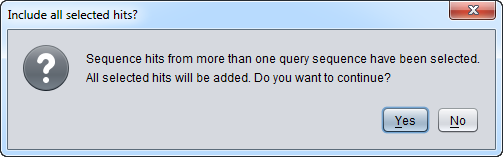
| + | ** '''''Note''''' - Starting with geWorkbench 2.4.0, sequence hits from more than one query sequence can be included in a single sequence set imported back into the [[Workspace|Workspace]]. Move between different result sets by selecting the desired query sequences one at a time at left in the results window, and then within each result set select the desired sequences. A warning will appear when this option is used, to insure that the user really intended to include results from multiple hits. | ||
| − | + | [[Image:BLAST_multiple_selected_warning.png]] | |
| − | * | + | ===At the bottom of the pane=== |
| + | * '''Reset''' - uncheck all "Include" boxes. | ||
| + | * '''Select All''' - mark as checked all the "Include" boxes. | ||
| + | * '''Add Complete Sequences to Workspace''' - for each hit whose "Include" box is checked, add its full sequence to a sequence node in the [[Workspace|Workspace]]. This option performs a retrieval query against the NCBI database to fetch the full sequence corresponding to each selected hit, not just the aligned portions. | ||
| + | ** Technical note - The query uses the integer Entrez database id (e.g. GI number) for a sequence, and this id will be reflected in the fasta format sequence entry returned as a sequence node to the Workspace. | ||
| + | * '''Only Add Aligned Parts''' - for each hit whose "Include" box is checked, add to the [[Workspace|Workspace]] only the portion(s) of its sequence which aligned with the query sequence. The new sequence node will contain one sequence for each aligned region. | ||
| + | ** Technical note - these sequences will be displayed with the accession number, as was the original hit. | ||
| + | *** The tag "---PARTIALLY INCLUDED" will also be appended to the accession number in the sequence node. | ||
| + | *** If there are sub-sequence in the hit, they will be indicated with (''n'') appended just after the accession number, where ''n'' is the number of the sub-sequence, e.g. "(1)---PARTIALLY INCLUDED". | ||
| − | * | + | ===In the query list=== |
| + | * '''Search''' - input a text search to find entries in the list of queries. | ||
| + | * '''Find Next''' - search for the next occurence of the entered text. | ||
| − | + | ==Adding selected sequence hits to the Workspace== | |
| + | The sequences corresponding to individual hits in the BLAST search can be retrieved from NCBI and added to the [[Workspace|Workspace]]. | ||
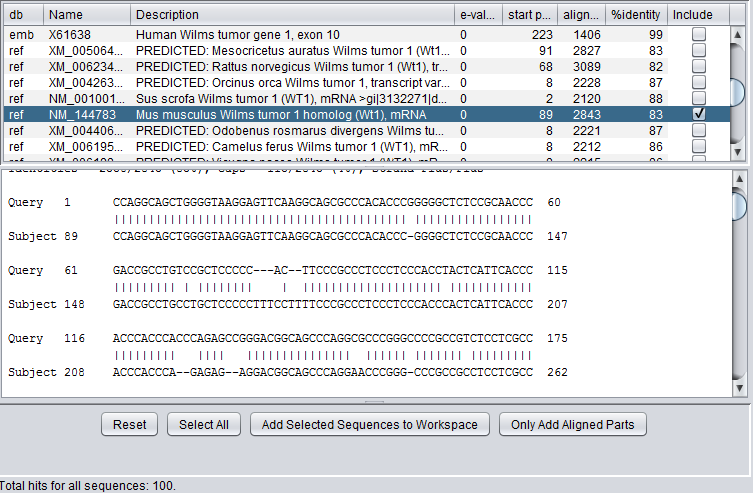
| − | + | [[Image:BLAST_Wilms_NM_select_mus.png|{{ImageMaxWidth}}]] | |
| − | [[ | + | Here we select a particular hit, by checking the "include" box next to it, and press the button "Add Selected Sequences to Workspace". The sequence is retrieved and placed into the [[Workspace|Workspace]] as shown below: |
| + | [[Image:BLAST_Wilms_NM_select_mus_added.png]] | ||
| − | |||
| + | ==Complete vs Aligned Sequences in the Workspace== | ||
| − | + | The Wilms tumor sequence was queried against the Human EST database. The final hit, with accession DB442323.1, shows the kind of difference that can occur between retrieving a complete sequence (first picture) and only the aligned parts of a sequence (second picture). | |
| − | |||
| + | Complete sequence for hit: | ||
| − | |||
| − | + | [[Image:BLAST_humEST_complete_seq.png]] | |
| − | |||
| − | + | Aligned part of hit only: | |
| − | |||
| − | [[Image: | + | [[Image:BLAST_humEST_aligned_only.png]] |
| + | =Example: Running a BLAST search= | ||
| + | Two Genbank sequence files in FASTA format are included in the geWorkbench data/public_data folder: a nucleotide sequence, "NM _024426-Wilms.Fasta", and its protein sequence, "NP_077744-Wilms.fasta". | ||
| − | + | For a simple search using the nucleotide query file, one can select the '''blastn/megablast''' program and search against the '''nr/nt''' database of nucleotide sequences. | |
| − | + | * In the [[Workspace|Workspace]], | |
| + | ** right-click on the [[Workspace|Workspace]] icon and select "Open File(s)", or | ||
| + | ** from the top-level "File" menu, select "Open->File". | ||
| + | * Select a file type of FASTA. | ||
| + | * Navigate to data/public_data within the geWorkbench distribution and select the file "NM _024426-Wilms.Fasta". | ||
| + | * Press "Open". | ||
| + | * Right-click on the new sequence data node. | ||
| + | * Select Analysis->BLAST Analysis. | ||
| − | |||
| − | + | * For program select '''blastn''', and leave the default algorithm choice set to megablast. | |
| + | * For database, select '''nr''' - the complete nucleotide database. | ||
| − | + | '''Note:''' - If you have a fasta file that has multiple sequences, you can create a set in the Markers component and use only this set for the search, by activating the set (check the box by its name in the Markers component). You can override an activated Marker Set and search on all sequences in a file by clicking the '''All Markers''' checkbox. | |
| + | * Click on the '''Algorithm Parameters''' Tab | ||
| + | * Change the '''Expect threshold''' to 0.01. This sets the cutoff for which BLAST hits will be displayed. | ||
| + | * Leave the '''Display result in your web browser''' checked. | ||
| + | * Hit the "Analyze" button. The job will be submitted and the results returned as shown in the sections above. | ||
| − | + | =Technical Notes= | |
| + | * The NCBI BLAST server may occasionally return an error when sequences are searched from geWorkbench. The problem appears to depend on the load on the NCBI BLAST server. | ||
| + | * When geWorkbench is asked to submit multiple sequences to the NCBI BLAST server, it will submit them one at a time and wait for the results before submitting the next sequence. This is done to simplify the subsequent parsing and display of the results. | ||
| + | * '''Metagenome databases''' - The Whole Genome Shotgun (WGS) databases at NCBI include the metagenome samples. The metagenomic, or "environmental" samples sequences are currently also available in the env_nt and env_nr (metagenomic) databases, but this may not always be true. The NCBI BLAST website now only supports searching metagenomic projects through the WGS database. However, geWorkbench does not directly support searching the metagenomic projects via the WGS database, and instead continues to provide direct query of the metagenomes via env_nt and env_nr. (Additional details in Mantis #3198, #3801). | ||
| + | * '''Only first 100 hits displayed''' - geWorkbench only displays the first 100 hits returned by BLAST. The complete set of hits is shown in the web browser if this option is checked on the Algorithm Parameters tab. | ||
| + | =References= | ||
| + | * Berman P, Zhang Z, Wolf YI, Koonin EV, Miller W. (2000) Winnowing sequences from a database search. J Comput Biol. 7(1-2):293-302. | ||
| − | [ | + | * The NCBI BLAST site provides a comprehensive [http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastDocs&DOC_TYPE=References list of references]. |
Latest revision as of 23:41, 26 January 2015
Contents
- 1 Overview
- 2 NCBI Documentation
- 3 BLAST job setup
- 4 BLAST Results Viewer
- 5 Example: Running a BLAST search
- 6 Technical Notes
- 7 References
Overview
"The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families" (quoted from the NCBI BLAST homepage).
geWorkbench submits BLAST jobs to the NCBI server. NCBI-supported sequence databases and search algorithms can be selected in the user interface. Since release 2.1.0, geWorkbench supports almost all BLAST setting options available through the NCBI web interface.
Please note that although, in geWorkbench, we have adopted the default settings for each BLAST algorithm as seen on the NCBI website, those settings are subject to change at any time by NCBI. Before submitting a BLAST job from geWorkbench, the user should verify that the parameter settings are appropriate for their query.
The BLAST analysis is available when a protein or DNA sequence is loaded and selected in the Workspace. The BLAST Analysis and BLAST Results Viewer must be loaded in the Component Configuration Manager.
Figure legend: BLAST Main Parameters. A nucleotide sequence has been loaded into the Workspace.
NCBI Documentation
geWorkbench serves as a interface to the NCBI BLAST server and implements the same options as the NCBI BLAST website. Detailed information about each option can be found on NCBI webpages, including:
Older help pages...
BLAST job setup
Prerequisites
- The BLAST Analysis and Viewer components must be loaded in the Component Configuration Manager.
- A protein or nucleotide sequence must be loaded in the Workspace.
Query sequences
BLAST accepts nucleotide or amino-acid query sequences in the FASTA format. A query file can contain one or multiple sequences. The file can be loaded from disk using the File Open command, or may have been placed into the Workspace by another component such as the Sequence Retriever, or add from the result of a previous BLAST run.
Invoking BLAST
BLAST is a normal geWorkbench analysis component and can be invoked either by right-clicking on a sequence node in the Workspace, or through the Commands entry in the Menu Bar at the top of the geWorkbench GUI.
Invoking BLAST via right-clicking on a sequence node in the Workspace:
Invoking BLAST via the Commands menu. The desired sequence node must be selected first in the Workspace:
Parameters - Main
In addition to the parameters shown in the previous image, BLASTX and TBLASTX add the Genetic Code option:
Algorithms
The user must make sure that the algorithm chosen matches the type of query sequence (protein or nucleotide) that has been loaded. Some of the algorithms translate a nucleotide query, a nucleotide database, or both into amino acid sequence before executing the query. Searching in the amino-acid space is more sensitive for certain types of query, as it ignores synonymous, non-functional changes in nucleotide sequence.
For protein query sequences:
- blastp - Compares an amino acid query sequence against a protein sequence database.
- tblastn - Compares a amino acid query sequence against a nucleotide database translated in all reading frames.
For nucleotide query sequences:
- blastn - three algorithms are available under the blastn choice, in order of decreasing similarity of the query to the target sequences, Each compares a nucleotide query sequence against a nucleotide sequence database:
- megablast - optimize for highly similar sequences.
- discontinuous megablast - optimize for more dissimilar sequences
- blastn - optimize for somewhat similar sequences.
- blastx - Compares a nucleotide query sequence translated in all reading frames against a protein sequence database.
- tblastx - Compares the 6-frame translations of a nucleotide query sequence against the 6-frame translations of a nucleotide sequence database.
Databases
Standard protein and nucleic acid databases maintained at NCBI are supported. The appropriate databases for the search algorithm chosen will be displayed. A window to the right, "Database Details", displays a summary of the database currently selected in the list.
For nucleic acids:
- Nucleotide Collection (nr/nt)
- Reference RNA seqeunces (refseq_rna)
- Reference genomic sequneces (refseq_genomic)
- NCBI Genomes (chromosome)
- Expressed Sequence Tags (est)
- Human subset of EST (est_human)
- Mouse subset of EST (est_mouse)
- Non-human, non-mouse ESTs (est_others)
- Genomic survey sequences (gss)
- High throughput genomic sequences (htgs)
- Patent sequences (pat)
- Protein Data Bank (pdb)
- Human ALU repeat elements (alu)
- Sequence tagged sites (dbsts)
- Whole genome shotgun contigs (wgs)
- Metagenomic Samples (env_nt)
- Transcriptome Shotgun Assembly (tsa_nt)
For proteins:
- Non-redundant protein sequences (nr)
- Reference proteins (refseq_protein)
- UniProtKB/Swiss-prot (swissprot)
- Patented protein sequences (pat)
- Protein Data Bank sequences (pdb)
- Metagenomic proteins (env_nr)
- Transcriptome Shotgun Assembly (tsa_nr)
Search Choice
These settings provide the ability to restrict the search in certain ways
- Exclude: exclude certain specialized database entries from the search.
- Models (XM/XP)
- Uncultured/environmental sequences
- Entrez Query - an Entrez query can be entered directly to restrict a search e.g. to a particular species. Please see NCBI BLAST help. An example from that page is "Mus musculus[organism] AND biomol_mrna[properties]". This limits the search to mouse mRNA entries in the database.
Genetic Code
Genetic code to be used in blastx (and tblastx) translation of the query.
Algorithm Parameters
General Parameters
- Max target sequences - maximum number of hits to return.
- Automatically adjust parameters for short input sequences.
- Expect threshold - Expected number of chance matches in a random model.
- Word size - The length of the seed that initiates an alignment.
- Max matches in a query range - Limit the number of matches to a query range. This option is useful if many strong matches to one part of a query may prevent BLAST from presenting weaker matches to another part of the query.
- Note - NCBI reports that this feature may not work if there are a large number of full-length sequence matches in the chosen database.
- The function of this feature is described in Berman et al., 2000.
Scoring Parameters
- Match/mismatch scores (blastn, megablast, discontinuous megablast) - scores to use for a match or mismatch.
- Matrix - Various scoring matrices (BLOSUM, PAM) are available for protein and translated queries.
- Gap Costs - The pull-down menu shows the available choices of gap costs for the current scoring matrix.
- Compositional adjustments - "...takes into account the amino acid composition of the individual database sequences involved in reported alignments. This improves E-value accuracy, thereby reducing the number of false positive results."
Filters and Masking
- Low Complexity - filter out low compositional complexity sequence.
- Species-specific repeats filter - masks species-specific repeats (e.g. human LINE's and SINE's). This option can speed searches involving long query sequences or databases containing sequences with many repeats.
- Mask for lookup table only - masks low-complexity sequence only while constructing the lookup table used by the initial hit-find phase of BLAST. The second phase, hit extension, is not not affected and hits can be extended through low-complexity sequence. NCBI notes that this option is experimental and subject to change.
- Mask lower case letter - filter out sequence which is in lower case in the FASTA query sequence.
Discontiguous Word Options
Please see the NCBI page on discontiguous megablast for a detailed explanation of these options.
- Template Length
- Template Type
Other
- Display result in your web browser - geWorkbench will display the HTML page returned by NCBI BLAST in your web browser as well is within its own display.
- Restore defaults - restore all settings for the currently selected algorithm to their default values.
Algorithm Parameter Setting Defaults
The default settings for each query type were taken from the NCBI BLAST website.
Please note that options and default settings on the NCBI BLAST website are subject to change at any time.
The user should verify all settings are appropriate for his or her particular BLAST query.
blastn - megablast
blastn - discontiguous megablast
blastn - blastn
blastp
blastx
tblastx
tblastn
Analyze
The BLAST analysis is launched by pushing the Analyze button. A dialog with a progress bar will appear. The analysis can be canceled by pushing the Cancel button on this dialog.
BLAST Results Viewer
When the BLAST search results are returned they are placed in a new node in the Workspace as a child of the query sequence used. Mousing over the result set will show how many sequences are in it.
Each different hit is listed on a line in the results table, shown below. Note that a query sequence can hit a database target sequence in more than one place, resulting in multiple alignments displayed per target hit. The results viewer also shows statisics for each hit, including the E-value, start position and length of the hit, and the percent identity.
If the "Display result in your web browser" option was chosen, then the browser will open with the HTML formated results.
In the pane at left in the picture below, the name of the input query sequence is shown, e.g. the gi number of a Genbank sequence. If there had been more than one query sequence, then this pane would show a list of query sequence names, allowing you to select the results to be viewed.
Controls
Within the list of returned hits
- Include check boxes - when checked, selects these sequences for import into the Workspace.
- Note - Starting with geWorkbench 2.4.0, sequence hits from more than one query sequence can be included in a single sequence set imported back into the Workspace. Move between different result sets by selecting the desired query sequences one at a time at left in the results window, and then within each result set select the desired sequences. A warning will appear when this option is used, to insure that the user really intended to include results from multiple hits.
At the bottom of the pane
- Reset - uncheck all "Include" boxes.
- Select All - mark as checked all the "Include" boxes.
- Add Complete Sequences to Workspace - for each hit whose "Include" box is checked, add its full sequence to a sequence node in the Workspace. This option performs a retrieval query against the NCBI database to fetch the full sequence corresponding to each selected hit, not just the aligned portions.
- Technical note - The query uses the integer Entrez database id (e.g. GI number) for a sequence, and this id will be reflected in the fasta format sequence entry returned as a sequence node to the Workspace.
- Only Add Aligned Parts - for each hit whose "Include" box is checked, add to the Workspace only the portion(s) of its sequence which aligned with the query sequence. The new sequence node will contain one sequence for each aligned region.
- Technical note - these sequences will be displayed with the accession number, as was the original hit.
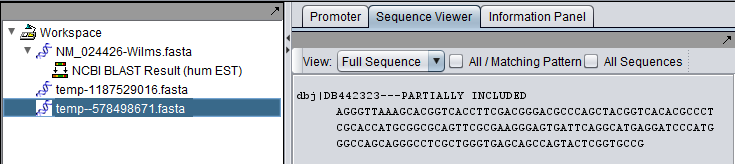
- The tag "---PARTIALLY INCLUDED" will also be appended to the accession number in the sequence node.
- If there are sub-sequence in the hit, they will be indicated with (n) appended just after the accession number, where n is the number of the sub-sequence, e.g. "(1)---PARTIALLY INCLUDED".
- Technical note - these sequences will be displayed with the accession number, as was the original hit.
In the query list
- Search - input a text search to find entries in the list of queries.
- Find Next - search for the next occurence of the entered text.
Adding selected sequence hits to the Workspace
The sequences corresponding to individual hits in the BLAST search can be retrieved from NCBI and added to the Workspace.
Here we select a particular hit, by checking the "include" box next to it, and press the button "Add Selected Sequences to Workspace". The sequence is retrieved and placed into the Workspace as shown below:
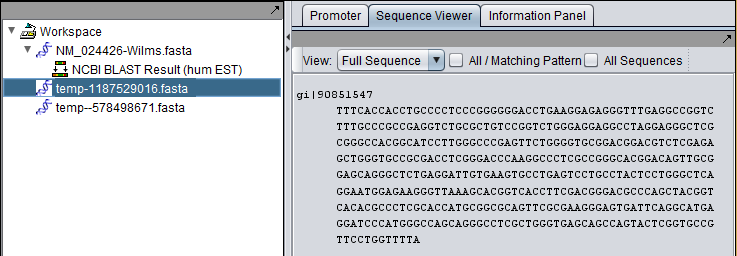
Complete vs Aligned Sequences in the Workspace
The Wilms tumor sequence was queried against the Human EST database. The final hit, with accession DB442323.1, shows the kind of difference that can occur between retrieving a complete sequence (first picture) and only the aligned parts of a sequence (second picture).
Complete sequence for hit:
Aligned part of hit only:
Example: Running a BLAST search
Two Genbank sequence files in FASTA format are included in the geWorkbench data/public_data folder: a nucleotide sequence, "NM _024426-Wilms.Fasta", and its protein sequence, "NP_077744-Wilms.fasta".
For a simple search using the nucleotide query file, one can select the blastn/megablast program and search against the nr/nt database of nucleotide sequences.
- In the Workspace,
- right-click on the Workspace icon and select "Open File(s)", or
- from the top-level "File" menu, select "Open->File".
- Select a file type of FASTA.
- Navigate to data/public_data within the geWorkbench distribution and select the file "NM _024426-Wilms.Fasta".
- Press "Open".
- Right-click on the new sequence data node.
- Select Analysis->BLAST Analysis.
- For program select blastn, and leave the default algorithm choice set to megablast.
- For database, select nr - the complete nucleotide database.
Note: - If you have a fasta file that has multiple sequences, you can create a set in the Markers component and use only this set for the search, by activating the set (check the box by its name in the Markers component). You can override an activated Marker Set and search on all sequences in a file by clicking the All Markers checkbox.
- Click on the Algorithm Parameters Tab
- Change the Expect threshold to 0.01. This sets the cutoff for which BLAST hits will be displayed.
- Leave the Display result in your web browser checked.
- Hit the "Analyze" button. The job will be submitted and the results returned as shown in the sections above.
Technical Notes
- The NCBI BLAST server may occasionally return an error when sequences are searched from geWorkbench. The problem appears to depend on the load on the NCBI BLAST server.
- When geWorkbench is asked to submit multiple sequences to the NCBI BLAST server, it will submit them one at a time and wait for the results before submitting the next sequence. This is done to simplify the subsequent parsing and display of the results.
- Metagenome databases - The Whole Genome Shotgun (WGS) databases at NCBI include the metagenome samples. The metagenomic, or "environmental" samples sequences are currently also available in the env_nt and env_nr (metagenomic) databases, but this may not always be true. The NCBI BLAST website now only supports searching metagenomic projects through the WGS database. However, geWorkbench does not directly support searching the metagenomic projects via the WGS database, and instead continues to provide direct query of the metagenomes via env_nt and env_nr. (Additional details in Mantis #3198, #3801).
- Only first 100 hits displayed - geWorkbench only displays the first 100 hits returned by BLAST. The complete set of hits is shown in the web browser if this option is checked on the Algorithm Parameters tab.
References
- Berman P, Zhang Z, Wolf YI, Koonin EV, Miller W. (2000) Winnowing sequences from a database search. J Comput Biol. 7(1-2):293-302.
- The NCBI BLAST site provides a comprehensive list of references.