Difference between revisions of "BLAST"
(→Overview) |
(→Databases) |
||
| Line 42: | Line 42: | ||
===Databases=== | ===Databases=== | ||
| − | Standard protein and nucleic acid databases maintained at NCBI are supported. | + | Standard protein and nucleic acid databases maintained at NCBI are supported. The appropriate databases for the search algorithm chosen will be displayed. |
For nucleic acids: | For nucleic acids: | ||
Revision as of 17:10, 30 September 2009
Contents
Outline
In this tutorial, we will:
- Discuss the various files, databases and algorithms involved in a BLAST search.
- Set up a search using a nucleotide sequence.
- Submit the job.
- View the returned results.
Overview
The BLAST algorithm is found in the Sequence Alignment tab, located in the command area of geWorkbench (lower right quadrant). The Sequence Alignment tab appears when a protein or DNA sequence is loaded and selected in the Project Folders component. BLAST is currently the only alignment option supported.
The BLAST algorithm is used to find similarities between nucleotide or amino acid query sequences and sequences held in a database. It is often used to give clues to the function of a sequence based on its similarity to already characterized sequences.
geWorkbench runs BLAST by submitting jobs to the NCBI server. There is no provision at this time for running a local BLAST job on the client desktop machine.
Query sequences
BLAST accepts nucleotide or amino-acid query sequences in the FASTA format. A query file can contain one or multiple sequences. The file can be loaded from disk using the File Open command, or may have been placed into the Project Folders component by another component such as the Sequence Retriever, or as a result of a previous BLAST run.
Paramaters - Main
Algorithms
There are five different query programs one can run:
blastp - Compares an amino acid query sequence against a protein sequence database.
blastn - Compares a nucleotide query sequence against a nucleotide sequence database.
blastx - Compares a nucleotide query sequence translated in all reading frames against a protein sequence database.
tblastn - Compares a amino acid query sequence against a nucleotide database dynamically translated in all reading frames.
tblastx - Compares the 6 frame translations of a nucleotide query sequence against the six frame translations of a nucleotide sequence database. This last is needless to say very time consuming!
It falls to the user to make sure the query sequence is of the appropriate type for the algorithm chosen.
Searches using translated sequences
As shown above, there are algorithm options which allow either a nucleotide query, a nucleotide database, or both to be searched after being translated into amino-acid sequences. Searching in the amino-acid space is more sensitive for certain types of query, as it ignores synonymous, non-functional changes in nucleotide sequence.
Databases
Standard protein and nucleic acid databases maintained at NCBI are supported. The appropriate databases for the search algorithm chosen will be displayed.
For nucleic acids:
- ncbi/nt - all non-redundant DNA sequences.
- ncbi/pdbnt - nucleotide sequences derived from the PDB database (protein 3D structure database).
- ncbi/yeast.nt - yeast genonic sequences.
For proteins:
- ncbi/nr - all non-redundant protein sequences
- ncbi/pdbaa - protein sequences from the PDB database (protein 3D structure database).
- ncbi/swissprot - sequences from Swiss-Prot, a primary reference database.
- ncbi/yeast.aa - translations of yeast genomic coding regions.
Setting up a search
Two Genbank Fasta sequence files are provided in the tutorial dataset, a nucleotide sequence, "NM _024426-Wilms.Fasta", and its protein sequence, "NP_077744-Wilms.fasta".
For a simple search using the nucleotide query file, one can select the blastn program and search against the ncbi/nt non-redundant database of nucleotide sequences. For an even quicker example search, one could run the protein query sequence against a small protein database derived from those sequences found in the PDB database of proteins having known structures.
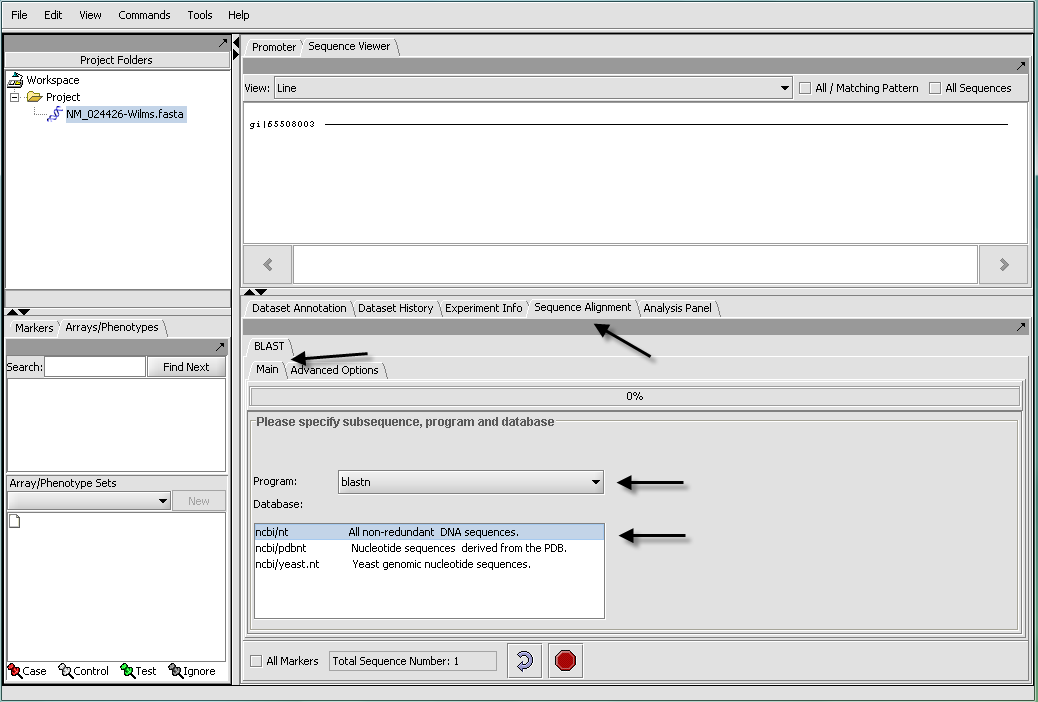
Here we will illustrate a search using the nucleotide file "NM _024426-Wilms.Fasta".
- Read the "NM _024426-Wilms.Fasta" data file into the Project component using the File Open command and file type FASTA.
- In the Project component, make sure the sequence file just read in is selected. This will activate those components that can work with sequence data.
- In the Commands Area click on the Sequence Alignment tab.
- Select the BLAST tab.
The length of the sequence is shown, and if desired a subset of the input sequence can be specified for use in the search. If more than one sequence was read in, the length of the longest is displayed.
- Click on the drop down arrow and select a program. Since this is a nucelotide query, here we select a nucleotide query program blastn.
- Select the desired nucleotide database. Here select ncbi/nt - the complete non-redundant nucleotide database. For a faster search, one could select the ncbi/pdbnt database instead, which is much smaller.
Note: The text field at the bottom of the Sequence Alignment component shows the number of sequences that have been selected. If you have a Fasta file that has multiple sequences, you can select the ones you want in the Markers component and activate this selection, letting you search on a subset. You can search on all sequences in a file by clicking the All Markers checkbox.
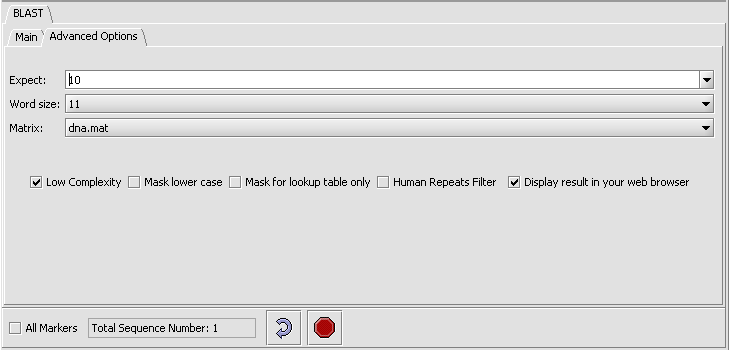
- Click on the Advanced Options Tab
- Change the Expect Value to 0.01. This sets the cutoff for which BLAST hits will be displayed.
- Make sure "dna mat" is selected for the Matrix.
- Leave the Display result in your web browser checked.
Submitting the BLAST job
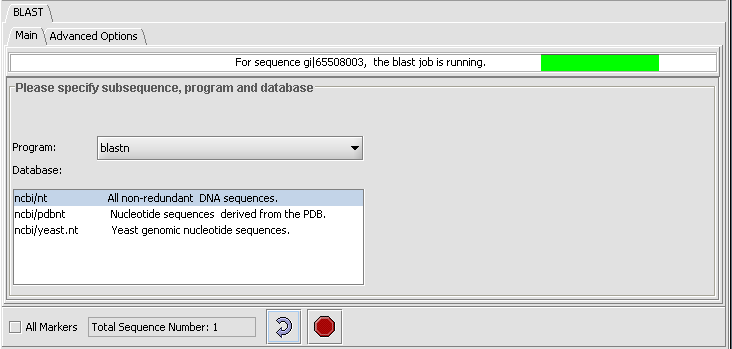
- Press the curved arrow submit button. The adjancent Stop button will terminate the search (geWorkbench will not wait for or retrieve the BLAST results).
- Returning to the Main tab; there the progress bar will show first the sequence being uploaded and then that the BLAST job is running.
Viewing the returned results
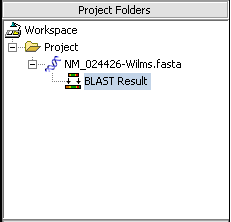
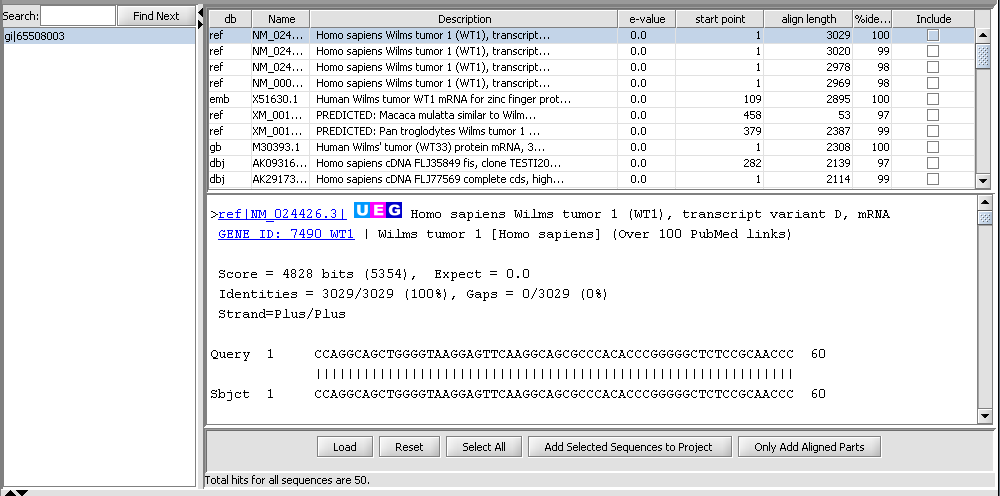
When the results are returned they are placed in the Project Folders as a child of the sequence they correspond to. You can mouse over the result set to see how many sequences are in it.
The result file will be opened in your web browser, if this option was selected. The alignment results are also displayed directly in the geWorkbench Blast results viewer and can be there further manipulated. Each different target hit is listed on a line in the results table. Note that a query sequence can hit a database target sequence in more than one place, resulting in mulitple alignments displayed per target hit.
In the Blast results viewer you can select sequences to add back to the main project by checking the include box and then the Add Selected Sequences To Your Project button.
You can also add just the aligned parts by clicking on the button Only Add Aligned Parts.
The results viewer also shows statisics for each hit, including the E-value, start and length of the hit, and the percent identity.
In the pane at left in the picture below, the name of the input query sequence is shown, e.g. the gi number of a Genbank sequence. Note that the "gi" designator is not shown, though it should be. If there had been more than one, then this would show a list of names, and you could select for which input query you wish to view the alignment results.
In the Blast results viewer, the Load button allows one to load an external Blast file in HTML format into the viewer.