BLAST
Overview
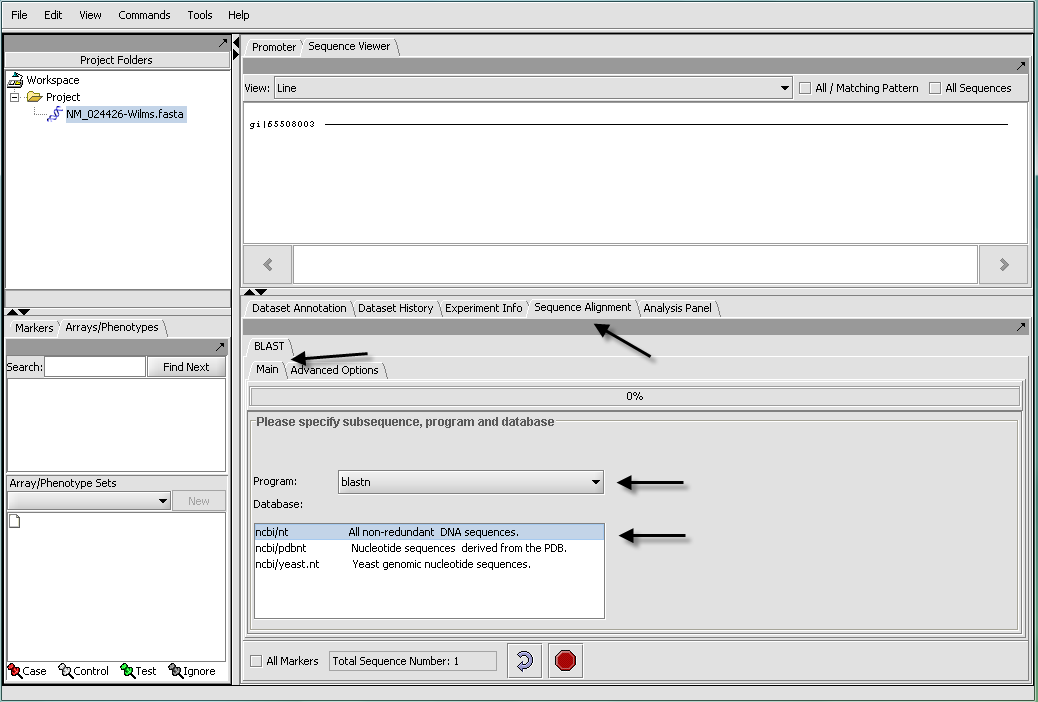
The BLAST algorithm is found in the Sequence Alignment tab, located in the command area of geWorkbench (lower right quadrant). The Sequence Alignment tab appears when a protein or DNA sequence is loaded and selected in the Project Folders component. BLAST is currently the only alignment option supported.
The BLAST algorithm is used to find similarities between nucleotide or amino acid query sequences and sequences held in a database. It is often used to give clues to the function of a sequence based on its similarity to already characterized sequences.
geWorkbench runs BLAST by submitting jobs to the NCBI server. NCBI-supported sequence databases and search algorithms can be selected in the user interface (arrows). There is no provision at this time for running a local BLAST job on the client desktop machine.
BLAST job setup
Prerequisites
- The Sequence Alignment component must be loaded in the geWorkbench Component Configuration Manager.
- A protein or nucleotide sequence must be loaded in the Project Folders component.
Query sequences
BLAST accepts nucleotide or amino-acid query sequences in the FASTA format. A query file can contain one or multiple sequences. The file can be loaded from disk using the File Open command, or may have been placed into the Project Folders component by another component such as the Sequence Retriever, or as a result of a previous BLAST run.
Parameters - Main
Algorithms
The user must make sure that the algorithm chosen matches the type of query sequence (protein or nucleotide) that has been loaded. Some of the algorithms translate a nucleotide query, a nucleotide database, or both into amino acid sequence before executing the query. Searching in the amino-acid space is more sensitive for certain types of query, as it ignores synonymous, non-functional changes in nucleotide sequence.
For protein query sequences:
- blastp - Compares an amino acid query sequence against a protein sequence database.
- tblastn - Compares a amino acid query sequence against a nucleotide database translated in all reading frames.
For nucleotide query sequences:
- blastn - Compares a nucleotide query sequence against a nucleotide sequence database.
- blastx - Compares a nucleotide query sequence translated in all reading frames against a protein sequence database.
- tblastx - Compares the 6 frame translations of a nucleotide query sequence against the six frame translations of a nucleotide sequence database.
Databases
Standard protein and nucleic acid databases maintained at NCBI are supported. The appropriate databases for the search algorithm chosen will be displayed.
For nucleic acids:
- ncbi/nt - all non-redundant DNA sequences.
- ncbi/pdbnt - nucleotide sequences derived from the PDB database (protein 3D structure database).
- ncbi/yeast.nt - yeast genonic sequences.
For proteins:
- ncbi/nr - all non-redundant protein sequences
- ncbi/pdbaa - protein sequences from the PDB database (protein 3D structure database).
- ncbi/swissprot - sequences from Swiss-Prot, a primary reference database.
- ncbi/yeast.aa - translations of yeast genomic coding regions.
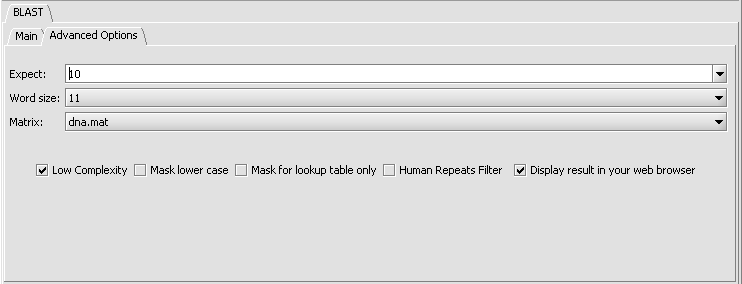
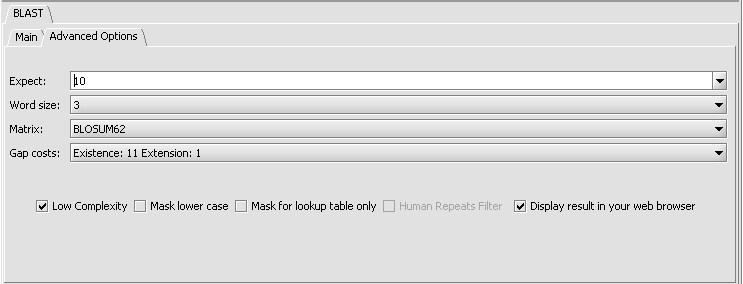
Parameters - Advanced Options
- Expect - Expected number of chance matches in a random model.
- Word size - The length of the seed that initiates an alignment.
- Matrix - Various scoring matrices are available for protein queries. A default matrix is used for DNA searches. The chosen matrix assigns a score for aligning any possible pair of residues. (A future release will include match/mismatch scores for DNA searches).
- Gap Costs - The pull-down menu shows the available choices of gap costs for the chosen matrix. Increasing the gap costs will tend to decrease the number of gaps in returned alignments. This is currently only implemented in geWorkbench for protein searches.
- Low Complexity - filter out low compositional complexity sequence.
- Mask lower case - filter out sequence which is in lower case in the FASTA query sequence.
- Mask for lookup table only - masks low-complexity sequence only while constructing the lookup table used by the intial hit-find phase of BLAST. The second phase, hit extension, is not not affected and hits can be extended through low-complexity sequence. NCBI notes that this option is experimental and subject to change.
- Human repeats filter - masks human repeats (LINE's and SINE's). This option can speed searches involving long query sequences or databases containing sequences with many repeats.
- Display result in your web browser - geWorkbench will display the HTML page returned by NCBI BLAST in your web browser as well is within its own display.
Please see the NCBI BLAST Help page 1 and NCBI BLAST help page 2 for further details on these options.
Nucleotide Search parameters (default):
Protein Search parameters (default):
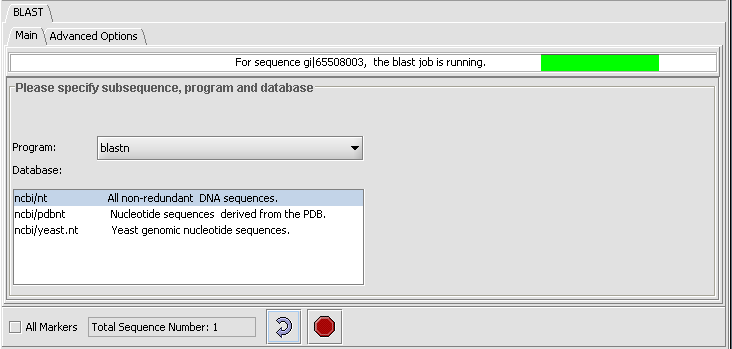
General controls
- All Markers - if selected, use all sequences loaded, overriding any activated sets in the Marker Sets component.
- Total Sequence Number - indicates how many sequences have been selected for query.
- Curling arrow - start BLAST search
- Stop sign - stop BLAST search (if pushed, geWorkbench will not wait for or retrieve the BLAST results).
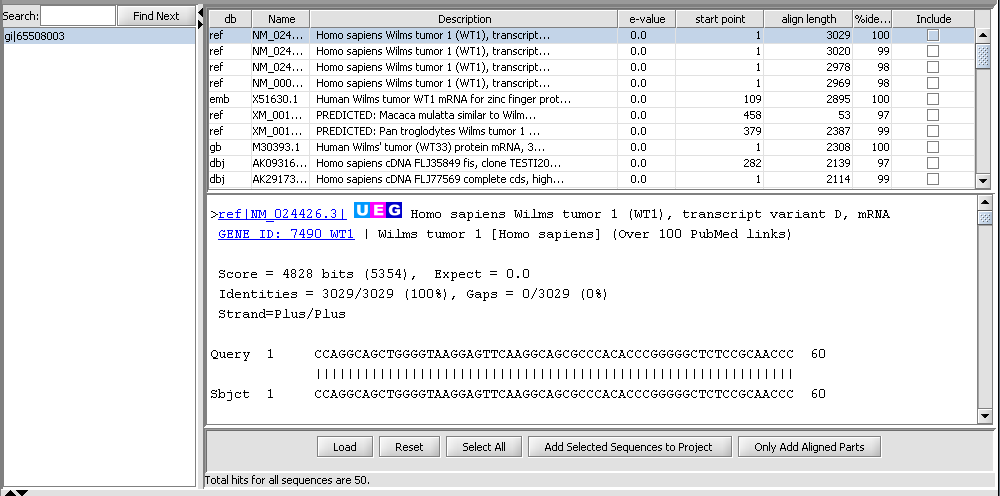
BLAST Results Viewer
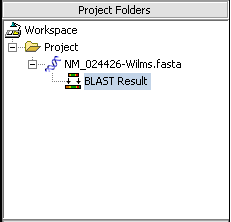
When the BLAST search results are returned they are placed in a new node in the Project Folders component as a child of the query sequence used. You can mouse over the result set to see how many sequences are in it.
Each different hit is listed on a line in the results table, shown below. Note that a query sequence can hit a database target sequence in more than one place, resulting in multiple alignments displayed per target hit. The results viewer also shows statisics for each hit, including the E-value, start position and length of the hit, and the percent identity.
If the "Display result in your web browser" option was chosen, then the browser will open with the HTML formated results.
In the pane at left in the picture below, the name of the input query sequence is shown, e.g. the gi number of a Genbank sequence. If there had been more than one query sequence, then this pane would show a list of query sequence names, allowing you to select the results to be viewed.
Controls
Within the list of returned hits
- Include check boxes - when checked, selects these sequences for import into the Project Folders component.
At the bottom of the pane
- Load - load a HTML format BLAST file into the viewer.
- Select All - mark as checked all the "Include" boxes.
- Reset - uncheck all "Include" boxes.
- Add Selected Sequences to Project - for each hit whose "Include" box is checked, add its sequence to a sequence node in the Project Folders component.
- Only Add Aligned Part - for each hit whose "Include" box is checked, add to the Project Folders component only the portion of its sequence which aligned with the query sequence .
In the query list
- Search - input a text search to find entries in the list of queries.
- Find Next - search for the next occurence of the entered text.
Submitting a BLAST job
- Press the curved arrow submit button. The adjacent Stop button will terminate the search (geWorkbench will not wait for or retrieve the BLAST results).
- Once the search has been submitted, a progress bar in the "Main" tab will indicate first that the sequence is being uploaded and then that the job is running.
Example: Running a BLAST search
Two Genbank sequence files in FASTA format are included in the geWorkbench data/public_data folder: a nucleotide sequence, "NM _024426-Wilms.Fasta", and its protein sequence, "NP_077744-Wilms.fasta".
For a simple search using the nucleotide query file, one can select the blastn program and search against the ncbi/nt non-redundant database of nucleotide sequences.
- From the geWorkbench "File" menu, select "Open->File".
- Select a file type of FASTA.
- Navigate to data/public_data within the geWorkbench distribution and select the file "NM _024426-Wilms.Fasta".
- Press "Open".
- (The above steps can also be accomplished by right-clicking on a Project node and selecting "Open File(s)" and following the same steps.
- In the Project Folders component, make sure the sequence file just read in is selected. This will activate those components that can work with sequence data.
- In the Commands Area click on the Sequence Alignment tab.
- Select the BLAST tab, and under it the Main tab.
- For program select blastn.
- For database, select "ncbi/nt - the complete non-redundant nucleotide database. For a faster search, one could select the ncbi/pdbnt database instead, which is much smaller.
Note: The text field at the bottom of the Sequence Alignment component shows the number of sequences that have been selected. If you have a Fasta file that has multiple sequences, you can select the ones you want in the Markers component and activate this selection, letting you search on a subset. You can search on all sequences in a file by clicking the All Markers checkbox.
- Click on the Advanced Options Tab
- Change the Expect Value to 0.01. This sets the cutoff for which BLAST hits will be displayed.
- Make sure "dna mat" is selected for the Matrix.
- Leave the Display result in your web browser checked.
- Hit the "curving arrow" run button. The job will be submitted and the results returned as shown in the sections above.