Difference between revisions of "Basics"
(→Window Decorations) |
|||
| Line 49: | Line 49: | ||
[[Image:Window_separator_handle.png]] | [[Image:Window_separator_handle.png]] | ||
| − | The small handle on each separator allows it to be positioned as desired by dragging. | + | The small handle on each separator allows it to be positioned as desired by dragging. |
| + | |||
| + | |||
| + | ==Command Popup== | ||
| + | |||
| + | Hitting F12 on the keyboard will pop-up a shortcut menu to the geWorkbench commands available for the current dataset. | ||
| + | |||
| + | [[Image:F12-Popup.png]] | ||
==Data management area== | ==Data management area== | ||
Revision as of 16:53, 13 March 2012
Contents
Layout of the Graphical User Interface
As of release 2.3.0
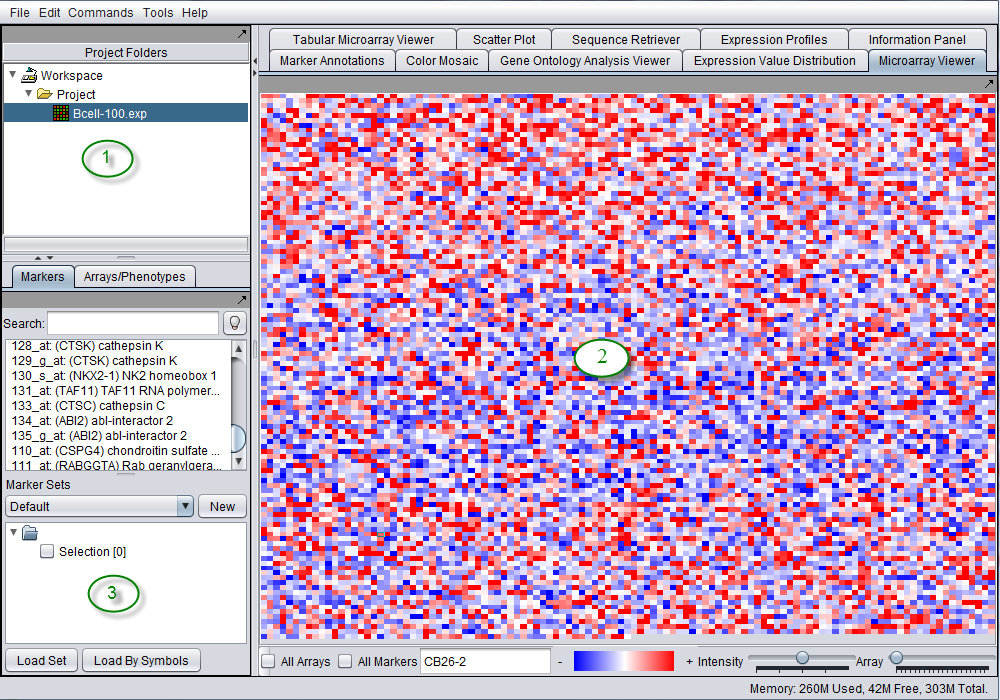
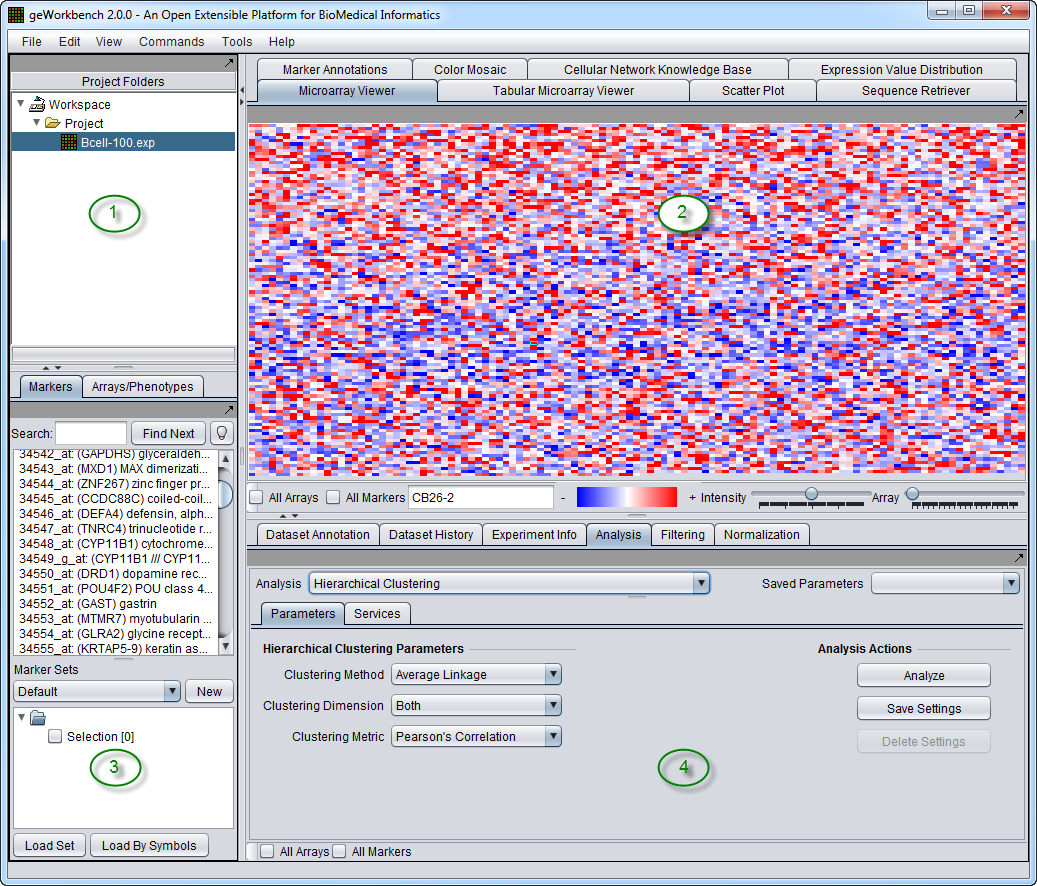
The graphical user interface for geWorkbench is divided into three major sections, for
- Data management - Workspace and Projects (upper left)
- Visualization tools (right)
- Marker and Array/Phenotype set selection and management (lower left)
Commands for analysis, filtering and normalization are now available in a menu activated by right-clicking on any loaded data node. They can also be found in the top level menu-bar under "Commands". They will act on the currently selected data node.
Prior to releases 2.3.0
The graphical user interface for geWorkbench is divided into four major sections, for
- Data management - Workspace and Projects (upper left)
- Visualization tools (upper right)
- Marker and Array/Phenotype set selection and management (lower left)
- Analytical tools (lower right)
Areas 2, 3 and 4 are defined for convenience. The actual placement of a given component into any of these three areas is controlled by a configuration file and can be customized as desired.
Window Decorations
The size and position of the various panes within the geWorkbench GUI can be easily adjusted.
Clicking on the upward-tilting arrow will undock a window from the GUI. It can then be expanded and positioned at will on the screen.
The two small left-right arrows allow the pane separator to be moved left or right all the way to the opposite edge.
The small handle on each separator allows it to be positioned as desired by dragging.
Command Popup
Hitting F12 on the keyboard will pop-up a shortcut menu to the geWorkbench commands available for the current dataset.
Data management area
Working with geWorkbench involves creating a project within the top-level workspace. Open data files and the results of data transformation or analysis are stored within a project. A workspace can contain more than one project at a time, allowing data to be organized as desired. A workspace and all the projects and data within it can be saved and later reloaded. These operations will be described in detail in further sections of the tutorials.
The GUI provides a menu bar at top with a standard choice of commands. Many commands that are available in the menu bar are also available by right-clicking on data objects.
Set selection and management
A key feature of geWorkbench is the ability to work with defined sets of markers or arrays. This allows subsets of data to be analyzed, and allows for passing of selected subsets of data between different components. For example, the t-test produces a list of markers showing a significant difference in expression between two states, and this list can then be used to retrieve relevant sequences or annotations.
Visualization and Analysis tools
geWorkbench works such that only the visualization and analysis components relevant to the type of dataset currently selected in the Project Folders area (1) are displayed through tabs in their respective areas (2 and 4). Thus choosing a microarray dataset will result in a different set of tabs being displayed as compared with those seen when a nucleotide sequence file is selected. When a new data file is loaded, or an analysis produces a new data set, not only is it added to the Project area (1), but an appropriate viewer in the Visualization area (2) is automatically selected.
Component Interoperability
The features described above underly the most important design goal of geWorkbench, which is to allow the different components to interoperate easily. The user is freed of the need to write programs to convert data from one format to another for different programs. Both the Project component (1) and the Set Selection component(3) can hold shared data. Typically each run of an analysis places either a new dataset into the active project, or a new set of markers or arrays into the Set Selection component. These are then available to any other appropriate component for reuse.
Data representation
geWorkbench works on a single dataset at one time, as selected in the Project Folders. For working with data from multiple microarrays simultaneously, for example for statistical analysis or clustering, all of the data must be represented as a single two-dimensional array of experiments and markers. Such a representation can be created externally, for example in Excel, or geWorkbench can read in expression results from single arrays and merge them into a combined dataset. The merging process is described in detail in the section Projects and Data Files.
An option at the time of file read-in allows multiple files to be opened and merged simultaneously, either from disk or from remote data sources such as caArray. Alternatively, single datasets that have already been read-in can also be merged at a later time. Either route creates a single data object in the project folder representing all of the arrays. This object can be viewed, for example, using the color mosaic component. The merged representation can be saved to the geWorkbench *.exp file type.
The results of any data analysis are stored in the Project Folders as a child of the dataset from which they were created.
Limitations
As currently implemented, operations such as filtering and normalization directly alter the loaded dataset. However, the original data file is not changed.