Color Mosaic
Contents
Overview
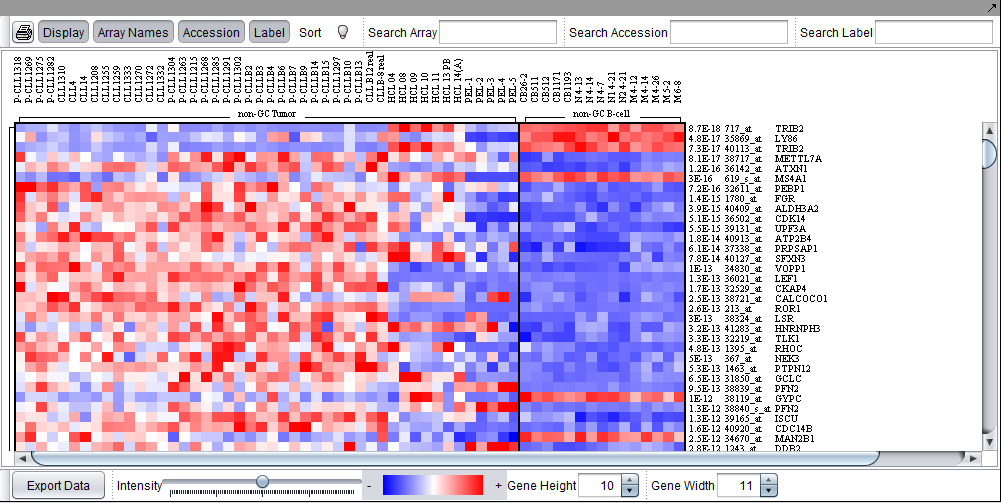
The Color Mosaic component displays expression values for all active arrays and markers using an "Eisen" plot "heat map" colored by either an absolute or relative color scheme. It is located in the View Area ( see figure below) in the top-right of the application.
Measurements in the selected microarray dataset are displayed as a two-dimensional matrix with each column representing an array and each row representing a gene/marker. The color spectrum is used to indicate the magnitudes of the measurements.
When the Color Mosaic component is used to display the results of a t-test or ANOVA, it will display the p-values to the right of each marker.
The figure below depicts the Color Mosaic component in "Relative" display mode. Three sets of arrays in the Bcell-100.exp tutorial dataset have been activated and each set is labeled in the display. The "Array Names", "Accession" and "Label" (gene name) buttons have been activated.
Controls
Display Controls
- Display - Toggles on/off the Color Mosaic display. It is better to leave the display "off" when not being used.
- Array Names - Toggles on/off the display of array names above the columns of the color mosaic.
- Accession - Toggles on/off the display of the accession number (e.g. Affymetrix probeset name) to the right of each row in the color mosaic.
- Label - Toggles on/off the display of the gene name (if available) to the right of each row in the color mosaic.
- Sort -
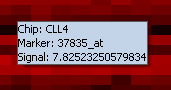
- Lightbulb - When activated, mouse-over (hover) text over any cell in the color mosaic will give the following information:
- Chip - the name of the array
- Marker - the marker name/accession number (e.g. Affymetrix probeset id).
- Signal - The value of the signal in the dataset for the marker/array pair.
- Intensity - The intensity slider increases or decreases the thresholds used to define the color mappings, as explained in the Color Key section.
- Gene Height and Gene Width - Change the size (number of pixels) assigned to each marker (row height) or array (column width) in the color mosaic. The associated labels scale proportionately.
Search Controls
Array names, accession numbers, and labels can be searched for using their respective search fields. The search fields are active only when the corresponding feature has been selected for display using the buttons described above. In each case, the next matching feature will be highlighted in blue. Hitting return again in the search box will advance the highlight to the next match.
- Search Array - Search array names.
- Search Accession - Search accession numbers.
- Search Label - Search marker names.
- Clear Search - clear the search result hightlights.
Other controls
- Export data - This option is only available when displaying results of an analysis, e.g. from ANOVA or t-test. Export p-values to a file.
- All Markers - Overrides any set of markers activated in the Markers component to display all markers. Disabled in some displays such as ANOVA.
- All Arrays - Overrides any set of arrays activated in the Arrays/Phenotypes component to display results from all arrays. Disabled in some displays such as ANOVA.
- Printer Icon - Send the heat-map to the printer.
Mouse-click actions
Left-clicking on an entry
Left-clicking on any location (tile) in the mosaic will highlight the corresponding gene probe in the Markers component, and the corresponding array in the Arrays/Phenotypes component.
Right-click
Right-clicking anywhere on the color mosaic display will bring up a menu with the following entries, as shown in the below figure:
- Print - Send the heat map to the printer.
- Zoom-in/ Zoom-out - Increase/decrease size of color mosaic.
- Take Snapshot - Save an image of the heat map to the Project Folders component.
- Export significance value - Export p-values from displayed ANOVA / t-test result.
Color key
The Color key shows the range of colors available in the currently set Visualization preference (Menu->Tools->Preferences->Visualization). There are two available color schemes: Absolute and Relative. In each, the lowest and highest expression values correspond to the most saturated colors, and values closer to the middle are indicated by shading into a third color.
Absolute
Colors are calculated on an absolute scale centered about zero (when the Intensity Slider is in the middle, neutral position). Values shade between the following:
- Black - zero.
- Red - positive values.
- Green - negative values.
Relative
Colors are calculated relative to the mean for each marker (when the Intensity Slider is in the middle, neutral position). Expression value colors are directly comparable only within a particular marker.
- White - mean value for marker.
- Red - values greater than the mean.
- Blue - values less than the mean.