Filtering
Contents
Overview
Filtering can be used to remove low quality data or reduce the size of the dataset by removing less interesting data. Most geWorkbench filters allow the user to specify a minimum number or percentage of arrays that must meet that filter's critereon before the marker will be removed.
Filter Configuration
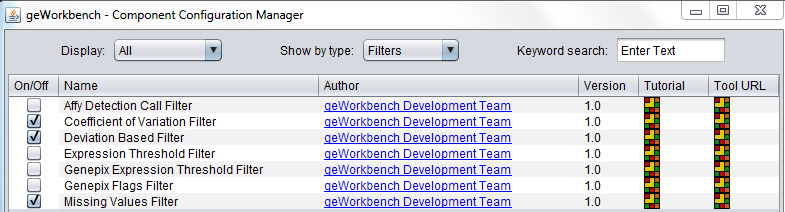
Some filters are not loaded by default in geWorkbench. To configure these filters, use the Component Configuration Manager (Tools->Component Configuration).
Available Filters
| Filter | Description |
|---|---|
| Affy Detection Call | Applicable to Affymetrix data only. Filter on Present, Marginal or Absent calls. |
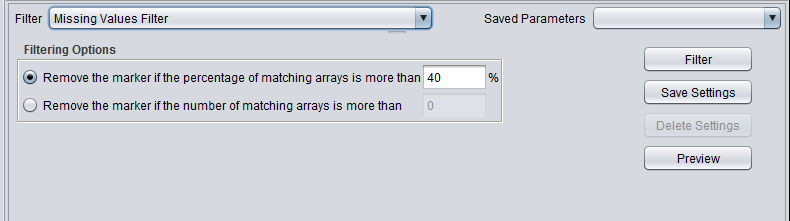
| Missing values | Removes markers that have “missing” measurements in more than a specified number (or percentage) of microarrays. |
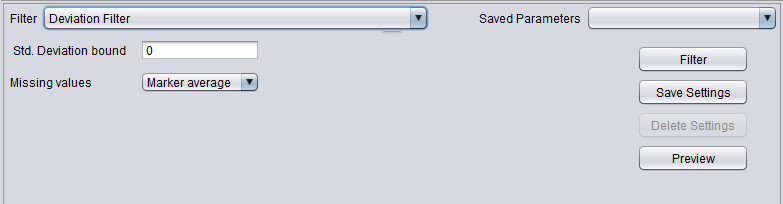
| Deviation | Removes markers whose standard deviation is less than a specified value across all microarrays. |
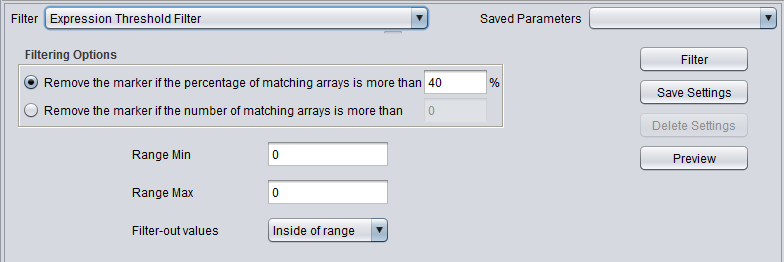
| Expression Threshold | Removes markers where more than a specified number (or percentage) have values inside (or outside) a user-defined range. |
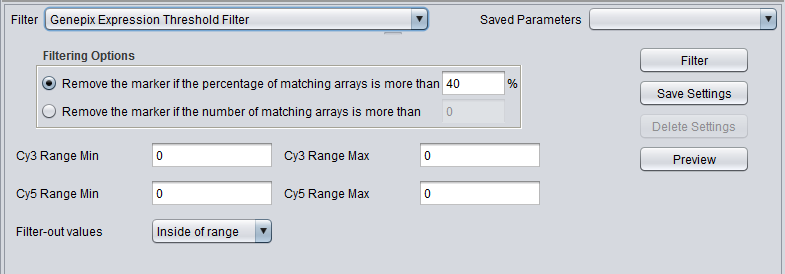
| Genepix Expression Threshold | Applicable to 2-channel arrays (Genepix) data only. Defines applicable ranges for each channel, and removes markers for which, for more than a specified number (or percentage) of markers, either channel intensity is inside (or outside) the defined range. |
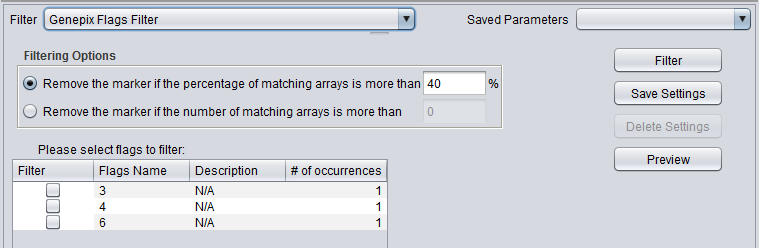
| GenePix Flags | Remove markers where more than a specified number (or percentage) of values match the selected flag (flagged in GenePix software). |
Basic Controls
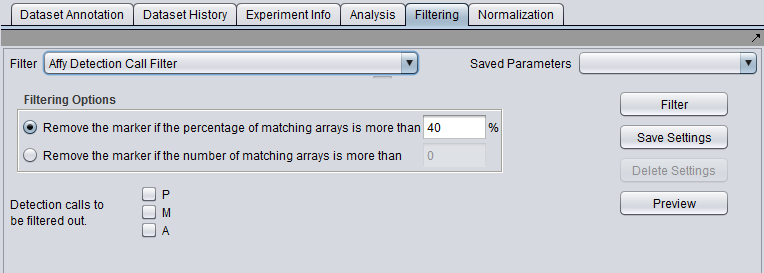
Filter - run the selected filter.
Preview - preview the filtering action (see following section).
Save Settings - Save the current settings (see following section).
Delete Settings- delete the currently selected parameter set.
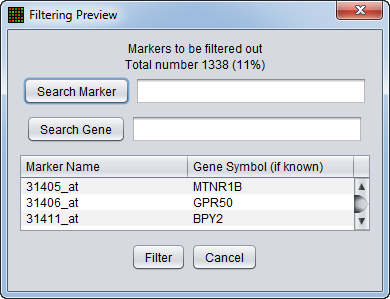
Preview
The filtering action can be previewed to allow the user to judge whether to proceed with the current parameter settings. The markers that will be removed are listed, as is a count of the markers in the list. The list displays marker names and, where available, gene names.
Search Marker - Search the list by marker name.
Search Gene - Search the list by gene name.
Filter - perform the filtering action.
Cancel - Cancel the filtering action, no change is made.
Saving Parameters
The current parameter settings can be saved to a named parameter set. The saved set will be displayed in the pull-down menu at upper right in the component. Any number of parameter sets can be saved. If the currently set parameters match a saved set, that set's entry will be shown in the menu.
Save Settings - save the current settings to a new parameter set.
Delete Settings - delete the currently selected parameter set from the menu.
Specific Controls for each Filter
Affymetrix Detection Call Filter
Deviation Filter
Expression Threshold Filter
GenePix Expression Threshold
GenePix Flag Filter