Difference between revisions of "IDEA"
(→Results) |
|||
| Line 14: | Line 14: | ||
=Results= | =Results= | ||
| − | == | + | ==Significant Genes tab== |
[[Image:IDEA_Significant_Genes.png]] | [[Image:IDEA_Significant_Genes.png]] | ||
| + | |||
| + | ==LOC Edges== | ||
| + | Loss of Correlation | ||
| + | |||
| + | |||
| + | ==GOC Edges== | ||
| + | Gain of Correlation | ||
Revision as of 16:39, 28 March 2012
Contents
IDEA
The IDEA component is being added to geWorkbench with release 2.3.0. Full documentation will be available in the near future. See Mani et al. 2008.
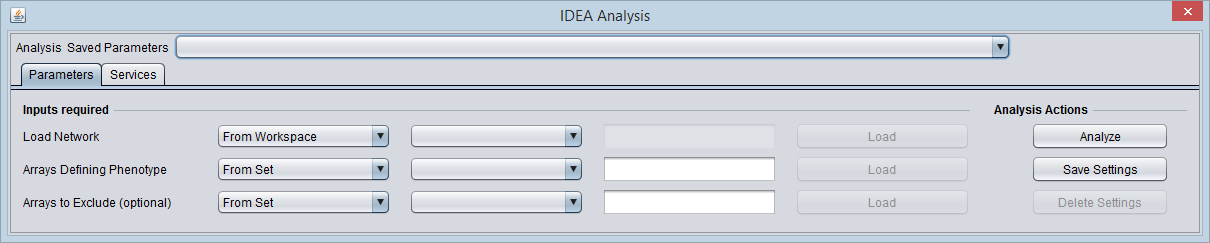
Analysis
Results
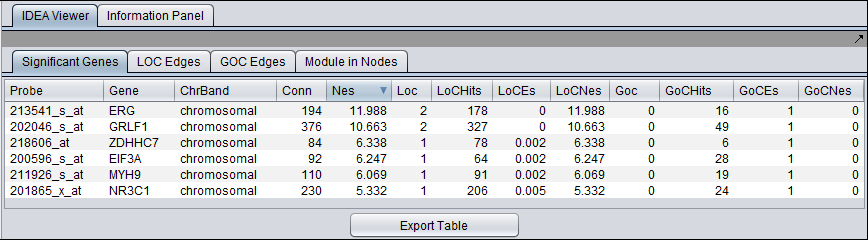
Significant Genes tab
LOC Edges
Loss of Correlation
GOC Edges
Gain of Correlation
Notes
Networks read in from files into the geWorkbench Project Folders component can be represented as Entrez ID, gene symbol, or probeset id. However, geWorkbench currently converts Entrez ID networks to gene symbols.
The IDEA analysis, for compatibility with its grid service implementation, first converts each network node, in case it is represented a gene symbol or probeset id, to an Entrez ID. In case multiple nodes map to the same Entrez ID, they are combined. In the case that a probeset is annotated to more than one Entrez ID, it is expanded to multiple nodes.
The network can be loaded from the Project, or loaded directly from file in ADJ or SIF format.
If the network contains nodes that are not found in the current microarray data set, a warning pop-up appears.
Network conversion to Entrez IDs is carried out after the "Analyze" button is pushed.
The mutual information parameter settings are apparently fixed, no preprocessing is available.
Only genes with a NES > 0 are included in the results table.
References
Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, Dalla-Favera R, Califano A. (2008) A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas. Mol Syst Biol. (4):169. PMID:18277385