Difference between revisions of "MarkUs"
(→Overview) |
|||
| Line 2: | Line 2: | ||
==Overview== | ==Overview== | ||
| − | + | ||
Mark-Us is a web server to assist the assessment of the biochemical function for a given protein structure. MarkUs identifies related protein structures and sequences, detects protein cavities, and calculates the surface electrostatic potentials and amino acid conservation profile. The results can be browsed by an interactive web interface that allows one to integrate Gene Ontology terms, UniProt features, and the Enzyme Classification. The Mark-Us website is at http://luna.bioc.columbia.edu/honiglab/mark-us/cgi-bin/submit.pl | Mark-Us is a web server to assist the assessment of the biochemical function for a given protein structure. MarkUs identifies related protein structures and sequences, detects protein cavities, and calculates the surface electrostatic potentials and amino acid conservation profile. The results can be browsed by an interactive web interface that allows one to integrate Gene Ontology terms, UniProt features, and the Enzyme Classification. The Mark-Us website is at http://luna.bioc.columbia.edu/honiglab/mark-us/cgi-bin/submit.pl | ||
Revision as of 12:11, 31 July 2009
Overview
Mark-Us is a web server to assist the assessment of the biochemical function for a given protein structure. MarkUs identifies related protein structures and sequences, detects protein cavities, and calculates the surface electrostatic potentials and amino acid conservation profile. The results can be browsed by an interactive web interface that allows one to integrate Gene Ontology terms, UniProt features, and the Enzyme Classification. The Mark-Us website is at http://luna.bioc.columbia.edu/honiglab/mark-us/cgi-bin/submit.pl
Mark-Us in geWorkbench
The starting point for a Mark-Us analysis is a PDB protein structure file.
1. If the Mark-Us component has not been loaded in the Component Configuration Manager (CCM), first do so.
2. Load a PDB file for the protein whose structure you would like to analyze.
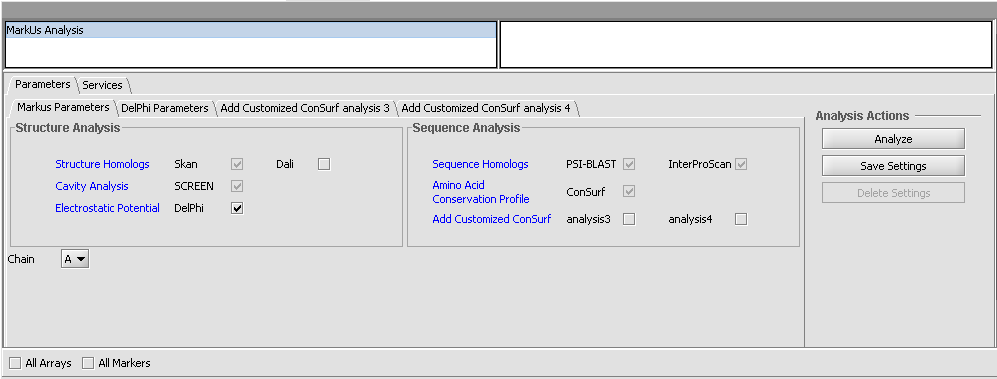
3. Set the desired parameters. The defaults are shown in the following figure.
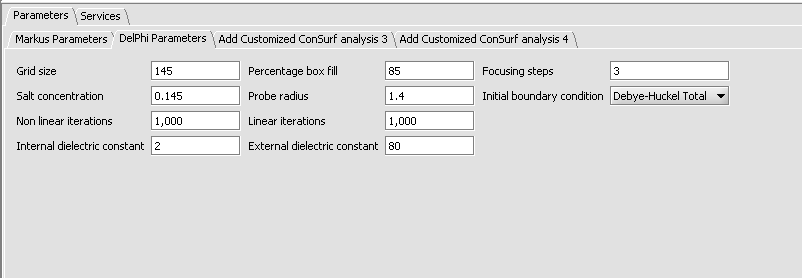
4. Inspect and adjust if desired the parameters to DelPhi. DelPhi calculates the electrostatic field around the molecule.
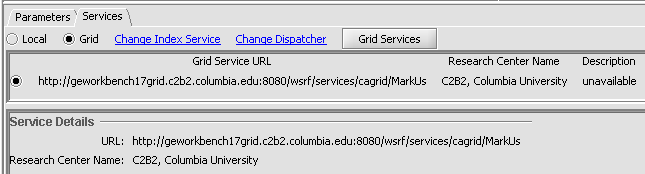
5. Mark-Us is available in geWorkbench only as a grid service.
- Click on the Services tab.
- Select the "Grid" radio button.
- Push the "Grid Services" button. This will retrieve available Mark-Us grid services from an index server.
- Select the radio button for the desired grid service.
6. Return to the Parameters tab.
7. Push the "Analyze" button.
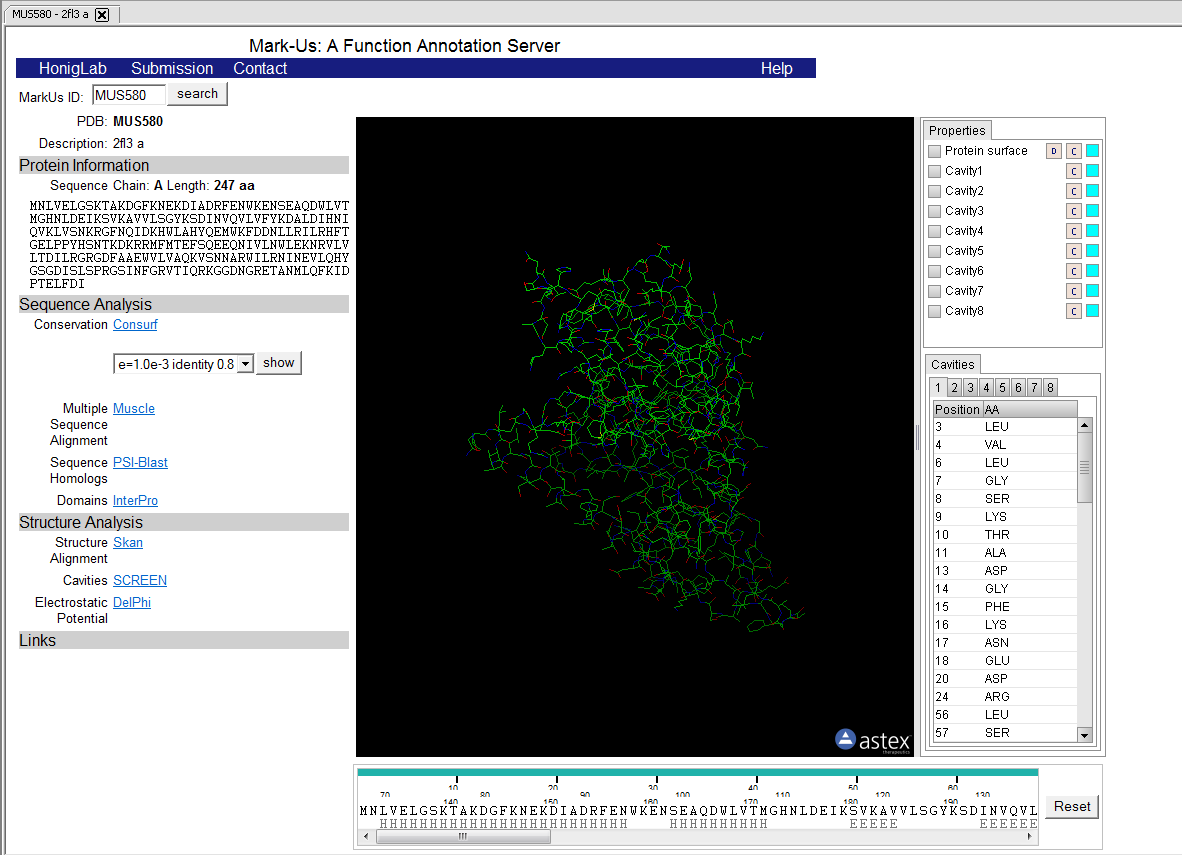
8. The result of the analysis is returned to a web-browser window displayed within geWorkbench.
Services (Grid)
Markus can only be run remotely as a grid job on caGrid. See the Grid Services section for further details on setting up a grid job.