Difference between revisions of "Master Regulator Analysis"
(→t-test) |
(→References) |
||
| (202 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
| − | + | =Overview= | |
| + | Regulatory activity in the context of specific cellular phenotypes can be investigated using interaction networks. These are graphs where nodes represent genes and an edge between two nodes A and B means that genes ''A'' and ''B'' are participants in the same regulatory activity. E.g., ''A'' can be a transcription factor for ''B''; or, ''A'' can be an miRNA that silences ''B''. Analysis of such regulatory networks [[#Basso2005 | [Basso et al., 2005]]] has convincingly demonstrated their scale-free nature which is dominated by a relatively small number of nodes with a large degree of connectivity. The genes corresponding to those nodes are known as "master regulators" and collectively orchestrate the regulatory program of the underlying cellular phenotype(s). | ||
| − | + | Master Regulator analysis [[#Lefebvre2010 | [Lefebvre et al., 2010]]] is an algorithm used to identify transcription factors whose targets (e.g., as represented in an ARACNe-generated interactome) are enriched for a particular gene signature (e.g. a list of differentially expressed genes). The enrichment is evaluated using a statistical test such as Fisher’s exact test or GSEA. The objective is to place the signature genes within a regulatory context and identify the master regulators responsible for coordinating their activity, thus highlighting the regulatory apparatus driving phenotypic differentiation. | |
| − | + | Specifically, given an interaction network ''I'', a (presumed) master regulator gene ''A'', and a set of signature genes, MRA computes the enrichment of the signature genes in the regulon of ''A'', where the regulon of ''A'' is defined as its neighbors in the interaction network ''I''. | |
| − | + | Interaction networks are represented as "adjacency matrices". An adjacency matrix lists the connections that each node takes part in, and includes a measure of the strength of that interaction (e.g. the mutual information in the case of matrices generated by ARACNe). | |
| − | + | Their are two master regulator analysis components implementing different methods to evaluate the enrichment of the signature in the regulon. Either method will quantify how likely it is to encounter an enrichment of (at least) the observed size by chance alone. A small p-value is taken to imply that gene ''A'' may play a significant role in mediating the regulatory program that leads to the differential phenotypes. | |
| − | |||
| − | |||
| − | |||
| − | == | + | * '''[[MRA-FET|FET Method (local service)]]''' - this method use Fisher's Exact Test. This method is implemented locally in geWorkbench. |
| + | * '''[[MARINa|MARINa Method *(grid service)]]''' - this method uses GSEA and differs in substantial ways from the FET-based method. This method is only implemented as a grid service and currently has restricted availability due to its computational cost. A t-test between two phenotype classes is built in to the implementation to produce the gene signature. | ||
| + | |||
| + | The MARINa method can use sample shuffling to correct for non-independance between the expression of various genes. Sample shuffling is not implemented for the MRA-FET method and hence in that method, the p-values are not directly comparable between genes. | ||
| + | |||
| + | Please note that MARINa does not employ any of the special gene lists available for use with the GSEA algorithm, such as [http://www.broadinstitute.org/gsea/msigdb/index.jsp. MSigDB]. It uses only a calculated list of differentially expressed genes and the regulon of the TF being tested. | ||
| + | |||
| + | With the release of geWorkbench 2.5.0, MRA-FET and MRA-MARINa are located in two separate sets of components, which can be loaded in the [[Component_Configuration_Manager| CCM]]. | ||
| + | |||
| + | =Setting up an MRA run= | ||
| + | |||
| + | ==Prerequisites== | ||
| + | * Either or both Master Regulator Analysis (MRA) components, MRA-FET and MRA-MARINa, must be loaded in the [[Component_Configuration_Manager | Component Configuration Manager]]. | ||
| + | |||
| + | [[Image:MRA-MARINa-CCM.png]] | ||
| + | |||
| + | |||
| + | ===Gene Expression dataset=== | ||
| + | A gene expression dataset in which the phenotypic signature was identified or can be demonstrated. A t-test of differential expression will be run to generate the graphic "bar code" display of the effect of the master regulator on its regulon or to generate the signature gene list (MARINa method). | ||
| + | |||
| + | ===Interaction Network=== | ||
| + | An interaction network in the form of an adjacency matrix (See [[File_Formats|File Formats]]. Networks can be loaded from a file, or calculated with ARACNe from a dataset which includes the particular cellular phenotypes being investigated. If calculating the network with ARACNe, all genes to be tested as possible master regulators should be used as hubs. | ||
| − | + | If the incorrect network format is chosen, the user is warned and the analysis setup is terminated. | |
| − | + | If the network is loaded into MARINa as gene symbols or Entrez IDs, it will be transformed (expanded) to include all probesets annotated to each such gene if an annotation file has been loaded for the expression dataset. | |
| − | + | ===Signature genes (FET method)=== | |
| − | + | A list of signature gene markers which distinguish between two phenotypes. This list may come from a t-test, clustering, or some combination of methods. The user must define this set using methods relevant to the particular dataset and study goals. | |
| − | === | + | ===Candidate master regulator list (FET method)=== |
| + | A set of gene markers that will be tested as candidate master regulators. This set may be comprised of e.g. transcription factor and signalling pathway genes. | ||
| − | + | ===Note on Marker Sets=== | |
| − | + | geWorkbench provides a mechanism to restrict some analyses to using certain sets of markers by "activating" these sets in the Markers component. However, as the MRA analysis component uses named marker sets directly, it does not respect the activation state of marker sets in the Markers component, and such activated sets will have no effect on the analysis. | |
| − | + | However, activating microarray sets would restrict the markers used in generating the "bar graph" by the MRA viewer. | |
| − | |||
| + | For this reason, no marker sets should be "activated" (their check-box checked) during MRA analysis. | ||
| + | ==Parameters and Settings== | ||
| + | ===Main=== | ||
| + | The settings on this tab apply to both the FET and MARINa methods. | ||
| − | |||
| + | ====Load Network==== | ||
| + | There are 2 ways to designate the interaction network, represented by an adjacency matrix, that will be used for computing the regulons of the candidate master regulator genes: | ||
| + | * '''From File''': by choosing a file that describes a network. | ||
| + | * '''From Workspace''': by selecting an adjacency matrix node from the [[Workspace|Workspace]] component. | ||
| + | =====Load Network from File===== | ||
| + | * The file loading controls will become active when this option is chosen. | ||
| + | * Press the "Load" button to bring up the file browser. | ||
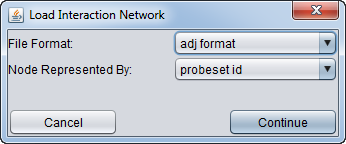
| + | * After selecting a file, a second dialog will ask for details about the format and symbols used. | ||
| − | + | [[Image:MRA_Load_Network_Dialog.png]] | |
| − | + | * '''File Format''': | |
| + | ** ADJ | ||
| + | ** SIF | ||
| + | ** MARINa 5-column format (internal use only) | ||
| − | + | * '''Nodes Represented by''': | |
| − | * | + | ** probeset id |
| − | * | + | ** gene symbol |
| − | ** | + | ** entrez id |
| − | ** | + | ** other |
| + | If the network is loaded into MARINa as gene symbols or Entrez ID, it will be transformed (expanded) to include all probesets annotated to each such gene if an annotation file has been loaded for the expression dataset. | ||
| − | + | After the file has been loaded, its name will be displayed in the adjacent text field. | |
| − | |||
| − | |||
| − | |||
| + | =====Load Network from Workspace===== | ||
| + | Several analytical components in geWorkbench (e.g., [[ARACNe | ARACNe]], [[Cellular_Networks_KnowledgeBase | CNKB]]) produce adjacency matrix results nodes that can be utilized for this purpose. Networks can also be loaded into the [[Workspace|Workspace]] directly from a file. | ||
| − | + | * The pulldown menu for choosing an available adjacency matrix will become active. Only adjacency matrices that are children of the current microarray dataset will be offered. | |
| − | |||
| − | |||
| − | |||
| − | + | All edges in the network are assumed to be significant, and any strength value included is not used. | |
| − | |||
| − | |||
| − | |||
| + | ====Enrichment Threshold==== | ||
| + | Enter a p-value for the significance at which to accept the overlap of the regulon of a candidate TF and the signature set of genes. | ||
| + | For the FET (local service), this is calculated using the FET. For the MARINa (grid service) method, this is calculated using GSEA. | ||
| − | [[ | + | ===MRA-FET (Local service)=== |
| + | Please see the separate [[MRA-FET|MRA-FET]] chapter for details on running the FET version of master regulator analysis. | ||
| − | == | + | ===MARINA (grid service)=== |
| − | + | Please see the separate [[MARINa|MARINa]] chapter for details on running MARINa. | |
| − | + | =Dataset History= | |
| + | Each results node stores the parameter settings used to setup the corresponding MRA run. The specific parameter values can be inspected within the Dataset History component, after clicking on the MRA results node in the [[Workspace]]. | ||
| − | == | + | =References= |
| − | + | <span id="Basso2005"></span> | |
| − | + | * Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A (2005). Reverse engineering of regulatory networks in human B cells. Nat Genet 37(4):382-390 ([http://www.nature.com/ng/journal/v37/n4/abs/ng1532.html link to paper]). | |
| − | + | * Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A (2010) The transcriptional network for mesenchymal transformation of brain tumors. Nature 463(7279):318-25. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/20032975 20032975]. | |
| − | + | <span id="Lefebvre2010"></span> | |
| − | + | * Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, Basso K, Beltrao P, Krogan N, Gautier J, Dalla-Favera R, Califano A (2010) A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 6:377. PMID: [http://www.ncbi.nlm.nih.gov/pubmed/20531406 20531406]. | |
| − | + | <span id="Lim2009"></span> | |
| + | * Lim WK, Lyashenko E, Califano A: Master regulators used as breast cancer metastasis classifier. Pac Symp Biocomput. 2009:504-15 ([http://psb.stanford.edu/psb-online/proceedings/psb09/lim.pdf link to paper]). | ||
| + | * Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9(3):157-73. | ||
Latest revision as of 17:47, 31 July 2014
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
Regulatory activity in the context of specific cellular phenotypes can be investigated using interaction networks. These are graphs where nodes represent genes and an edge between two nodes A and B means that genes A and B are participants in the same regulatory activity. E.g., A can be a transcription factor for B; or, A can be an miRNA that silences B. Analysis of such regulatory networks [Basso et al., 2005] has convincingly demonstrated their scale-free nature which is dominated by a relatively small number of nodes with a large degree of connectivity. The genes corresponding to those nodes are known as "master regulators" and collectively orchestrate the regulatory program of the underlying cellular phenotype(s).
Master Regulator analysis [Lefebvre et al., 2010] is an algorithm used to identify transcription factors whose targets (e.g., as represented in an ARACNe-generated interactome) are enriched for a particular gene signature (e.g. a list of differentially expressed genes). The enrichment is evaluated using a statistical test such as Fisher’s exact test or GSEA. The objective is to place the signature genes within a regulatory context and identify the master regulators responsible for coordinating their activity, thus highlighting the regulatory apparatus driving phenotypic differentiation.
Specifically, given an interaction network I, a (presumed) master regulator gene A, and a set of signature genes, MRA computes the enrichment of the signature genes in the regulon of A, where the regulon of A is defined as its neighbors in the interaction network I.
Interaction networks are represented as "adjacency matrices". An adjacency matrix lists the connections that each node takes part in, and includes a measure of the strength of that interaction (e.g. the mutual information in the case of matrices generated by ARACNe).
Their are two master regulator analysis components implementing different methods to evaluate the enrichment of the signature in the regulon. Either method will quantify how likely it is to encounter an enrichment of (at least) the observed size by chance alone. A small p-value is taken to imply that gene A may play a significant role in mediating the regulatory program that leads to the differential phenotypes.
- FET Method (local service) - this method use Fisher's Exact Test. This method is implemented locally in geWorkbench.
- MARINa Method *(grid service) - this method uses GSEA and differs in substantial ways from the FET-based method. This method is only implemented as a grid service and currently has restricted availability due to its computational cost. A t-test between two phenotype classes is built in to the implementation to produce the gene signature.
The MARINa method can use sample shuffling to correct for non-independance between the expression of various genes. Sample shuffling is not implemented for the MRA-FET method and hence in that method, the p-values are not directly comparable between genes.
Please note that MARINa does not employ any of the special gene lists available for use with the GSEA algorithm, such as MSigDB. It uses only a calculated list of differentially expressed genes and the regulon of the TF being tested.
With the release of geWorkbench 2.5.0, MRA-FET and MRA-MARINa are located in two separate sets of components, which can be loaded in the CCM.
Setting up an MRA run
Prerequisites
- Either or both Master Regulator Analysis (MRA) components, MRA-FET and MRA-MARINa, must be loaded in the Component Configuration Manager.
Gene Expression dataset
A gene expression dataset in which the phenotypic signature was identified or can be demonstrated. A t-test of differential expression will be run to generate the graphic "bar code" display of the effect of the master regulator on its regulon or to generate the signature gene list (MARINa method).
Interaction Network
An interaction network in the form of an adjacency matrix (See File Formats. Networks can be loaded from a file, or calculated with ARACNe from a dataset which includes the particular cellular phenotypes being investigated. If calculating the network with ARACNe, all genes to be tested as possible master regulators should be used as hubs.
If the incorrect network format is chosen, the user is warned and the analysis setup is terminated.
If the network is loaded into MARINa as gene symbols or Entrez IDs, it will be transformed (expanded) to include all probesets annotated to each such gene if an annotation file has been loaded for the expression dataset.
Signature genes (FET method)
A list of signature gene markers which distinguish between two phenotypes. This list may come from a t-test, clustering, or some combination of methods. The user must define this set using methods relevant to the particular dataset and study goals.
Candidate master regulator list (FET method)
A set of gene markers that will be tested as candidate master regulators. This set may be comprised of e.g. transcription factor and signalling pathway genes.
Note on Marker Sets
geWorkbench provides a mechanism to restrict some analyses to using certain sets of markers by "activating" these sets in the Markers component. However, as the MRA analysis component uses named marker sets directly, it does not respect the activation state of marker sets in the Markers component, and such activated sets will have no effect on the analysis.
However, activating microarray sets would restrict the markers used in generating the "bar graph" by the MRA viewer.
For this reason, no marker sets should be "activated" (their check-box checked) during MRA analysis.
Parameters and Settings
Main
The settings on this tab apply to both the FET and MARINa methods.
Load Network
There are 2 ways to designate the interaction network, represented by an adjacency matrix, that will be used for computing the regulons of the candidate master regulator genes:
- From File: by choosing a file that describes a network.
- From Workspace: by selecting an adjacency matrix node from the Workspace component.
Load Network from File
- The file loading controls will become active when this option is chosen.
- Press the "Load" button to bring up the file browser.
- After selecting a file, a second dialog will ask for details about the format and symbols used.
- File Format:
- ADJ
- SIF
- MARINa 5-column format (internal use only)
- Nodes Represented by:
- probeset id
- gene symbol
- entrez id
- other
If the network is loaded into MARINa as gene symbols or Entrez ID, it will be transformed (expanded) to include all probesets annotated to each such gene if an annotation file has been loaded for the expression dataset.
After the file has been loaded, its name will be displayed in the adjacent text field.
Load Network from Workspace
Several analytical components in geWorkbench (e.g., ARACNe, CNKB) produce adjacency matrix results nodes that can be utilized for this purpose. Networks can also be loaded into the Workspace directly from a file.
- The pulldown menu for choosing an available adjacency matrix will become active. Only adjacency matrices that are children of the current microarray dataset will be offered.
All edges in the network are assumed to be significant, and any strength value included is not used.
Enrichment Threshold
Enter a p-value for the significance at which to accept the overlap of the regulon of a candidate TF and the signature set of genes. For the FET (local service), this is calculated using the FET. For the MARINa (grid service) method, this is calculated using GSEA.
MRA-FET (Local service)
Please see the separate MRA-FET chapter for details on running the FET version of master regulator analysis.
MARINA (grid service)
Please see the separate MARINa chapter for details on running MARINa.
Dataset History
Each results node stores the parameter settings used to setup the corresponding MRA run. The specific parameter values can be inspected within the Dataset History component, after clicking on the MRA results node in the Workspace.
References
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A (2005). Reverse engineering of regulatory networks in human B cells. Nat Genet 37(4):382-390 (link to paper).
- Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A (2010) The transcriptional network for mesenchymal transformation of brain tumors. Nature 463(7279):318-25. PMID: 20032975.
- Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, Basso K, Beltrao P, Krogan N, Gautier J, Dalla-Favera R, Califano A (2010) A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 6:377. PMID: 20531406.
- Lim WK, Lyashenko E, Califano A: Master regulators used as breast cancer metastasis classifier. Pac Symp Biocomput. 2009:504-15 (link to paper).
- Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9(3):157-73.