Difference between revisions of "Master Regulator Analysis"
(→Running MRA) |
(→Overview) |
||
| Line 4: | Line 4: | ||
==Overview== | ==Overview== | ||
| − | The Master Regulator Analysis (MRA) component works with | + | The Master Regulator Analysis (MRA) component works with microarray gene expression data. |
| − | Its goal is to identify transcription factors (TFs) which control the regulation of a set of target genes (TGs) that demonstrate significant differential expression. Differential expression is measured using a simple t-test across 2 cellular phenotypes, e.g. “Case” and “Control”. | + | Its goal is to identify transcription factors (TFs) which control the regulation of a set of target genes (TGs) that demonstrate significant differential expression. Differential expression is measured using a simple t-test across 2 cellular phenotypes, e.g. “Case” and “Control”. Sets of genes controlled by TFs (the interaction network) are identified using a separate run of ARACNe. However, the dataset from which the interaction network is derived would not necessarily be the same one used for the t-test. An ARACNe run requires a dataset which explores many different expression phenotypes of a particular cell type, whereas a differential expression experiment compares only two classes. |
| − | The types of data which will be used are: | + | The types of data which will be used in the MRA then are: |
| − | # | + | # A microarray dataset appropriate for examining differential gene expression using a t-test. |
# A list of putative transcription factors which are to be tested against the differentially expressed genes. | # A list of putative transcription factors which are to be tested against the differentially expressed genes. | ||
| − | # An interaction network in the form of an ARACNe adjacency matrix. It should contain the results of an ARACNe run including, as hub markers, at least all of the transcription factors listed above | + | # An interaction network in the form of an ARACNe adjacency matrix. It should contain the results of an ARACNe run including, as hub markers, at least all of the transcription factors listed above. |
Revision as of 10:10, 15 July 2009
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
The Master Regulator Analysis (MRA) component works with microarray gene expression data. Its goal is to identify transcription factors (TFs) which control the regulation of a set of target genes (TGs) that demonstrate significant differential expression. Differential expression is measured using a simple t-test across 2 cellular phenotypes, e.g. “Case” and “Control”. Sets of genes controlled by TFs (the interaction network) are identified using a separate run of ARACNe. However, the dataset from which the interaction network is derived would not necessarily be the same one used for the t-test. An ARACNe run requires a dataset which explores many different expression phenotypes of a particular cell type, whereas a differential expression experiment compares only two classes.
The types of data which will be used in the MRA then are:
- A microarray dataset appropriate for examining differential gene expression using a t-test.
- A list of putative transcription factors which are to be tested against the differentially expressed genes.
- An interaction network in the form of an ARACNe adjacency matrix. It should contain the results of an ARACNe run including, as hub markers, at least all of the transcription factors listed above.
Briefly, the steps which MRA will execute are:
- Run a t-test on the the microarray dataset selected in the Project Folders component.
- For each TF, the set of its nearest neighbors (in the adjacency matrix), that is, those markers showing the closest interaction with it, are tested for overlap with the set of differentially expressed genes using Fisher's Exact test.
- The result is a list of transcription factors whose interactions show a significant overlap with the differentially expressed genes.
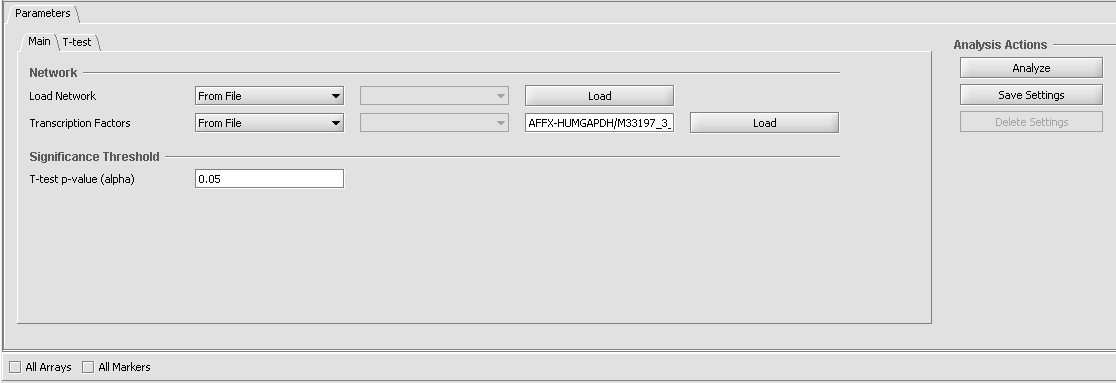
Parameters and Settings
Load Network
The network consists of an adjacency matrix generated by ARACNe.
- From File - load an adjacency matrix generated by an external run of ARACNe.
- From Project - load an ARACNe adjacency matrix from a result node in the Project Folders component.
Transcription Factors
- From File - Load a comma-separate list of transcription factors from a file.
- From Sets - Use a set defined in the Markers component as the list of transcription factors.
Significance Treshold
- T-test p-value (alpha) - The cutoff p-value by which to establish whether a particular marker shows a significant difference in expression between the two groups. (Note that multiple testing corrections are offered on the t-test parameters tab).
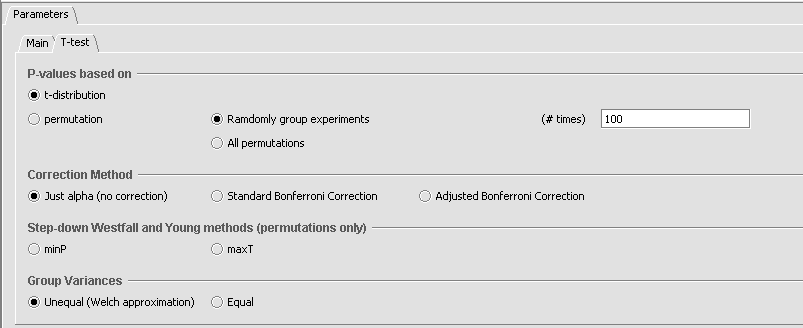
The parameter settings for the MRA t-test are shown in the figure below. These parameters are the same as those described in the t-test component tutorial
Running MRA
- Once all the parameters described above are set as desired, press the Analyze button. The t-test followed by the Fisher's Exact tests will be carried out.
- A table and graphic showing transcription factors for whose interactions significant overlap with the set of differentially expressed genes was found will be displayed.