Master Regulator Analysis
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
Regulatory activity in the context of specific cellular phenotypes can be investigated using interaction networks. These are graphs where nodes represent genes and an edge between two nodes A and B means that genes A and B are participants in the same regulatory activity. E.g., A can be a transcription factor for B; or, A can be an miRNA that silences B. Analysis of such regulatory networks [Basso et al., 2005] has convincingly demonstrated their scale-free nature which is dominated by a relatively small number of nodes with a large degree of connectivity. The genes corresponding to those nodes are known as "master regulators" and collectively orchestrate the regulatory program of the underlying cellular phenotype(s).
Master Regulator analysis [Lefebvre et al., 2010] is an algorithm used to identify transcription factors whose targets (e.g., as represented in an ARACNe-generated interactome) are enriched for a particular gene signature. The enrichment is evaluated using a statistical test such as Fisher’s exact test. The objective is to place the signature genes within a regulatory context and identify the master regulators responsible for coordinating their activity, thus highlighting the regulatory apparatus driving phenotypic differentiation.
Specifically, given an interaction network I, a (presumed) master regulator gene A, and a set of signature genes, MRA computes the intersection between two sets of genes:
- The neighbors of A in the interaction network I (this gene set is called the regulon of A).
- The set of signature genes. The signature may be supplied independently or calculated from e.g. a differential expression experiment.
Interaction networks are represented as "adjacency matrices". An adjacency matrix lists the connections that each node takes part in, and includes a measure of the strength of that interaction (e.g. the mutual information in the case of matrices generated by ARACNe).
Fisher’s exact test is used to quantify how likely it is to encounter an intersection of (at least) the observed size by chance alone. A small p-value is taken to imply that gene A may play a significant role in mediating the regulatory program that leads to the differential phenotypes.
Generating a display of the effect of a master regulator on its regulon requires performing a t-test of differential expression on a dataset representing the two phenotypes distinguished by the signature, ideally the original dataset from which the signature was determined.
Setting up an MRA run
Prerequisites
- The Master Regulator Analysis (MRA) component must be loaded in the Component Configuration Manager.
- The MRA component will be listed along with the other analysis routines within the geWorkbench Analysis Panel.
- Interaction Network - An interaction network calculated with ARACNe from a dataset which includes the particular cellular phenotypes being investigated. In calculating the network, all genes that will be tested as possible master regulators should be used as hubs in the ARACNe calculation.
- Signature genes - A list of signature genes which distinguish between two phenotypes. This list may come from a t-test, clustering, or some combination of methods. The user must define this set using methods relevant to the particular dataset and study goals.
- Candidate master regulator list - A set of genes that will be tested as candidate master regulators. This set may be comprised of e.g. transcription factor and signalling pathway genes.
- Gene Expression dataset - A gene expression dataset in which the phenotypic signature was identified or can be demonstrated. A t-test of differential expression will be run to generate the graphic "bar code" display of the effect of the master regulator on its regulon.
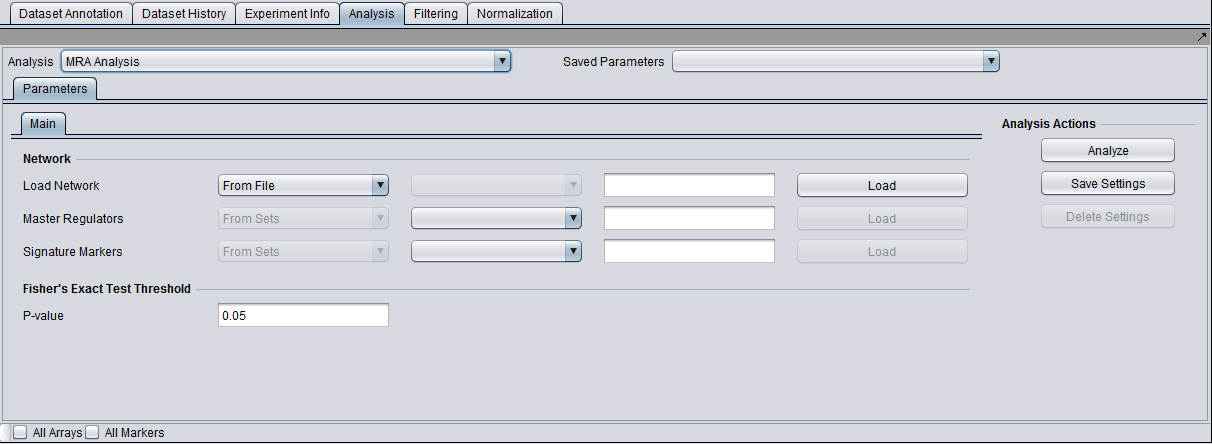
Parameters and Settings
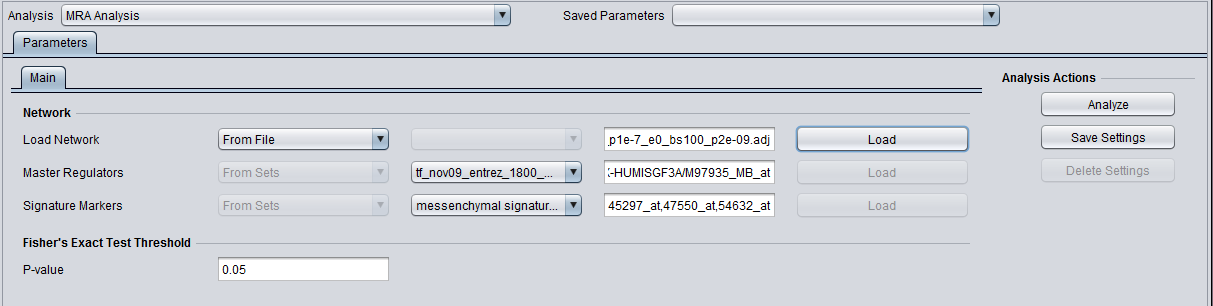
Load Network
There are 2 ways to designate the interaction network, represented by an adjacency matrix, that will be used for computing the regulons of the candidate master regulator genes:
- From File: by choosing a file that describes a network.
- From Project: by selecting an adjacency matrix node from the Project Folders component. Several analytical components in geWorkbench (e.g., ARACNE) produce adjacency matrix results nodes that can be utilized for this purpose.
All edges in the network are assumed to be significant, and any strength value included is not used.
Master Regulators
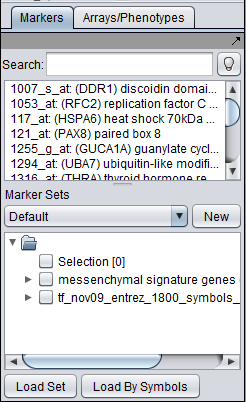
A set of candidate master regulator markers. This set must be loaded into the Markers component before running MRA. This can be read in either directly as markers, or as gene symbols.
Signature Markers
A set of markers comprising the signature that distinguishes the chosen phenotype from others. This set must be loaded into the Markers component before running MRA. It can be read in either directly as markers, or as gene symbols (but be aware that a gene can be represented by more than one marker, and not all may correspond to the signature).
T-test p-value (alpha)
Differential expression between the 2 phenotypes of interest is assessed using a t-test. The p-value provided by the user indicates the significance threshold below which a gene’s average expression is presumed to be significantly different in the 2 sets of arrays (cases and controls). Additional parameter settings affecting the execution of the t-test are defined within the “T-test” subtab. The parameters specified there are exactly the same as those used for the differential expression t-test analysis (see description in the corresponding tutorial).
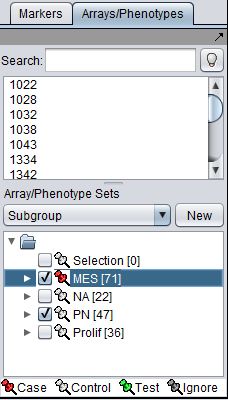
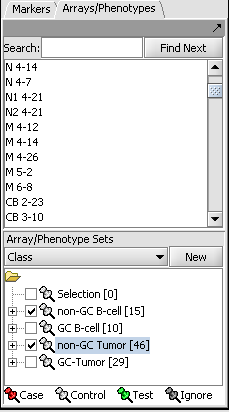
- Two array sets (each containing a distinct subset of arrays from the expression experiment) have been selected in the "Arrays/Phenotypes" component, representing the two phenotypes under investigation. One of the array sets (identified by the red-color pin) has been classified as containing the "Case" arrays (i.e., one of the 2 phenotypes) while the other one contains the "Controls" (the second phenotype):
NOTE:The "Case"/"Control" assignment is needed because, in general, it is possible to select more than one array sets for this analysis. In that case it is necessary to explicitly designate which belong to each of the 2 phenotypes.
Working with and viewing the analysis results
Following the successful completion of the MRA computation, a result node appears in the Project Folder area of the geWorkbench interface, under the microarray experiment node used for the computation:
The results of the analysis can be visualized in the MRA Viewer component by selecting the result node.
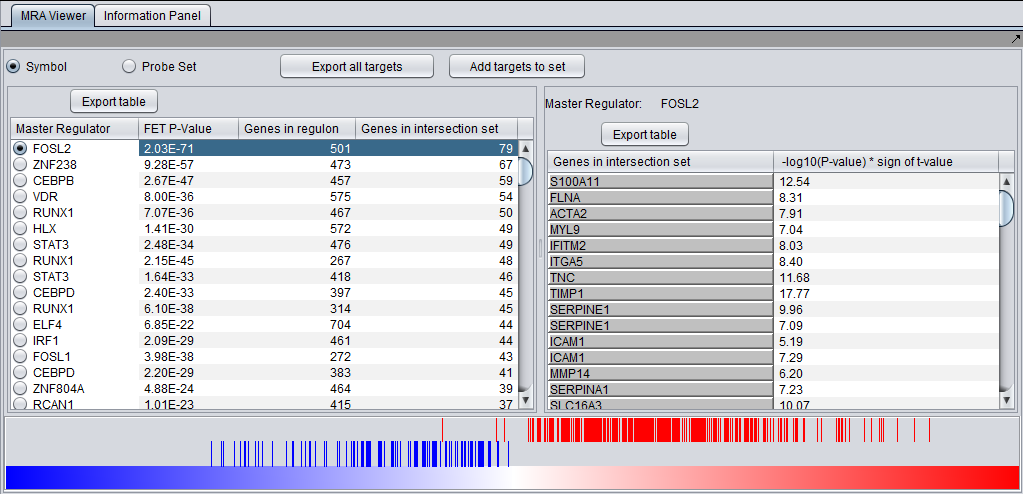
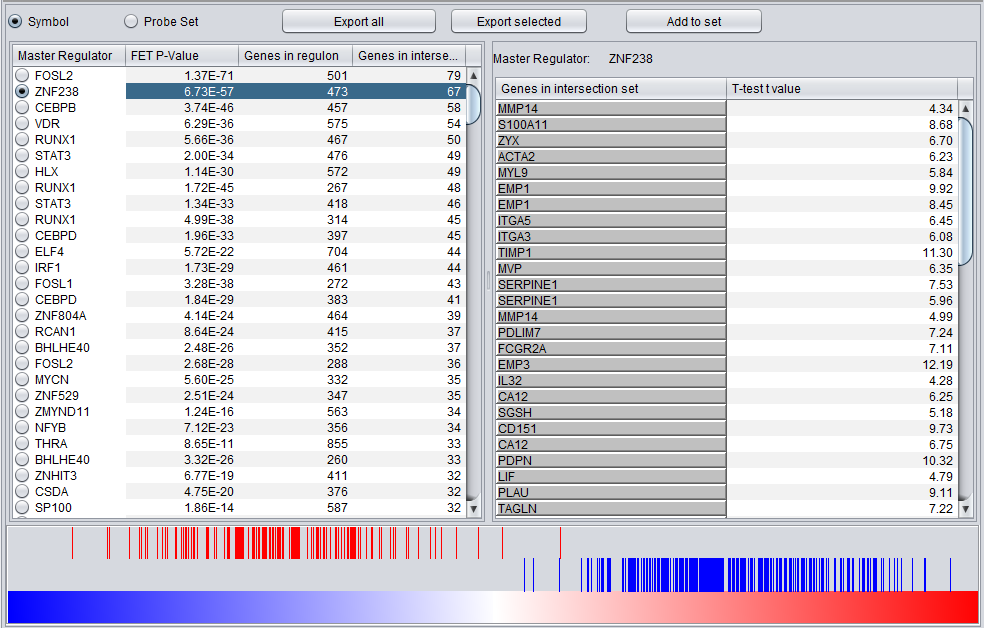
MRA Results Viewer
The MRA viewer is structured in 3 distinct areas.
FOSL2
ZNF238
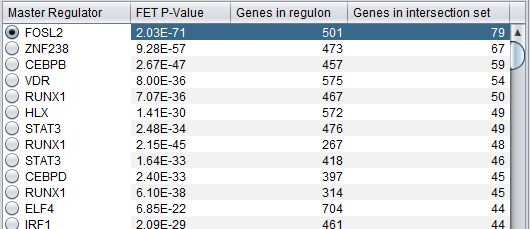
Summary Listing
This is a table with one row for each candidate master regulator.
Each row contains 4 columns:
- Master Regulator: This is either the master regulator gene name or the marker/probeset name identifying the corresponding array feature (depending on the selection of the radio buttons “Symbol” and “Probe set”).
- P-value: the p-value from Fisher’s exact test. The test utilizes a 2x2 contingency table where rows classify markers as differentially expressed versus non-differentially expressed, while columns indicate if a marker belongs to the regulon of the master regulator or not. Counts are computed using all markers found in the input experiment data. (Fischer's exact test includes p-values for more-extreme tables).
- Genes in regulon: The number of genes in the regulon of the master regulator (this comprises the set of genes that are first neighbors of the master regulator in the interaction network specified in the parameters panel).
- Genes in target list: The number of differentially expressed genes that are also members of the regulon.
The contents of the table can be ordered by any column, by clicking on the column name.
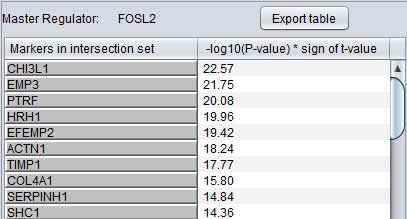
Detailed Listing
By clicking on the radio button associated with a master regulator in the Summary table, it is possible to display the complete listing of the differentially expressed genes intersecting the regulon of that master regulator.
The genes are displayed in a table with the following columns:
- Genes in target list: the names of the genes in the intersection set. Either the gene name or the marker/probe set name is used (based on the choice of "Symbol" or "Probe Set" radio buttons).
- P-value: the p-value of the t-test statistic for this marker, computed over the 2 sets of arrays (cases versus controls).
- T-test value: The actual value of the t-test statistic for the gene. A positive value indicates that the mean expression of the gene is higher in cases than in controls (a negative value has the opposite meaning).
The “P-val Threshold” parameter (located above the table) can be used to limit the number of target genes displayed. Specifically, only genes with a p-value less than the specified cutoff will be shown (if “P-val Threshold” is left empty, then all target genes in the intersection set are displayed). Additional functionality is made available through the following buttons:
- Add to Set: creates a set containing the markers that correspond to the genes currently displayed in the table. The new marker set appears in the Markers component and is named after the master regulator.
- Export selected: same as “Add to Set” but instead of being added to a marker set, the markers are stored into a file.
- Export all: stores into a file information for all master regulators (instead of only the one currently selected in the “Summary” view). Specifically, for each master regulator, the file lists the markers for all genes that are both differential expressed and also belong to the master regulator’s regulon.
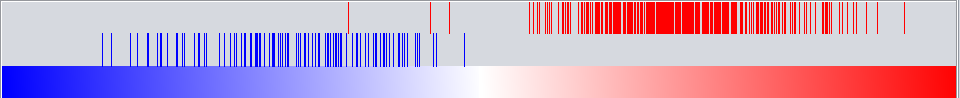
Graph View
For a given master regulator A and the intersection between its regulon and the set of differentially expressed genes, the graph view helps assess if the intersection genes are preferentially over-expressed in the cases versus the controls. The biological motivation comes from observing [Lim et al., 2009] that regulators with multiple targets tend to affect the expression level of (most of) their targets in one particular direction: they either promote their expression or inhibit it; but they rarely do both equally.
- The red-blue gradient at the bottom of the graph represents the range between the highest (red) and the lowest (blue) t-test statistic recorded among all differentially expressed genes.
- The vertical bars correspond to the genes displayed in the table under the “Detailed listing” portion of the interface, i.e., the intersection between the differentially expressed genes and the regulon of the master regulator A currently selected within the “Summary listing” table.
- The relative location of a bar on the gradient represents the t-test statistic recorded for the corresponding gene.
- Further, the color of each bar provides information about the correlation between the expression levels of the target gene and the master regulator A (correlations are computed as Pearson’s r, using data from all microarrays in the experiment): black means that the two genes are positively correlated (r > 0) while orange means that correlation is negative (r < 0).
Dataset History
Each results node stores the parameter settings used to setup the corresponding MRA run. The specific parameter values can be inspected within the Dataset History component, after clicking on the MRA results node in the Project Folders pane.
Example of running MRA
This example uses the Bcell-100.exp dataset available in the data/public_data directory of geWorkbench, and further described on the Download page. Briefly, this dataset is composed of 100 Affymetrix HG-U95Av2 arrays on which various B-cell lines, both normal and cancerous, were analyzed. Thus it explores a potentially wide variety of expression phenotypes.
Prerequisites
- Obtain the annotation file for the HG-U95Av2 array type from the Affymetrix NetAffx website (http://www.affymetrix.com/support/technical/byproduct.affx?product=hgu95). The name will be similar to "HG_U95Av2.na29.annot.csv", where na29 is the version number. Loading the annotation file associates gene names and other information with the Affymetrix probeset IDs (see the geWorkbench FAQ for details on obtaining these files).
- Store in your disk the following 2 files:
- Interaction_network.txt: describes an interaction network with 1955 nodes and 3810 edges. The file has 1955 lines (one for each node, the node name is the first entry in every line) and each line lists the edges emanating from that node. Edges are describes as tab-delimited pairs where the first member of the pair is the name of the target node and the second member is a number specifying a weight for the edge (MRA does not use the weight information but other geWorkbench components do). Node names correspond to marker ids.
- Master_regulators.csv: a list of master regulators. The marker ids in this file correspond to genes whose Gene Ontology annotation (under the Molecular Function category) lists them as transcription factors.
Loading and preparing the example data
- Load a microarray dataset. (See Local Data Files).
- When prompted, load the annotation file.
- Load marker sets for the list of candidate master regulators and for the signature genes.
Choosing array groups
If needed, define array sets for 2 classes of arrays in the dataset.
- Here we show the MES and PN sets selected.
- The MES set is classifed as "Case". Right click on the thumbtack adjacent to the set name.
Setting up the parameters and starting MRA
In the Analysis Panel, select the "MRA Analysis" entry and set the parameters as follows:
- Load Network: from the drop down choose the option "From Set", click on the "Load" button, and select the desired set.
- Transcription Factors: from the drop down choose the option "From Set", click on the "Load" button, and select the desired set.
- Click on the Analyze button.
Results
Upon completion of the analysis, an MRA results node is placed in the Project Folders tree. The analysis results can be browsed using the MRA viewer and are as shown above in the MRA Results Viewer section.
References
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A: Reverse engineering of regulatory networks in human B cells. Nat Genet 2005, 37(4):382-390 (link to paper).
- Lefebvre C, Rajbhandari P, Alvarez MJ, Bandaru P, Lim WK, Sato M, Wang K, Sumazin P, Kustagi M, Bisikirska BC, Basso K, Beltrao P, Krogan N, Gautier J, Dalla-Favera R, Califano A (2010) A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers. Mol Syst Biol. 6:377. PMID: 20531406 (link to paper).
- Lim WK, Lyashenko E, Califano A: Master regulators used as breast cancer metastasis classifier. Pac Symp Biocomput. 2009:504-15 (link to paper).