Microarray Dataset Viewers
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
In the this tutorial, you will:
- Get acquainted with the various geWorkbench visualization tools
- View a dataset in geWorkbench
- Modify the visualization preference settings
Visualization Tools
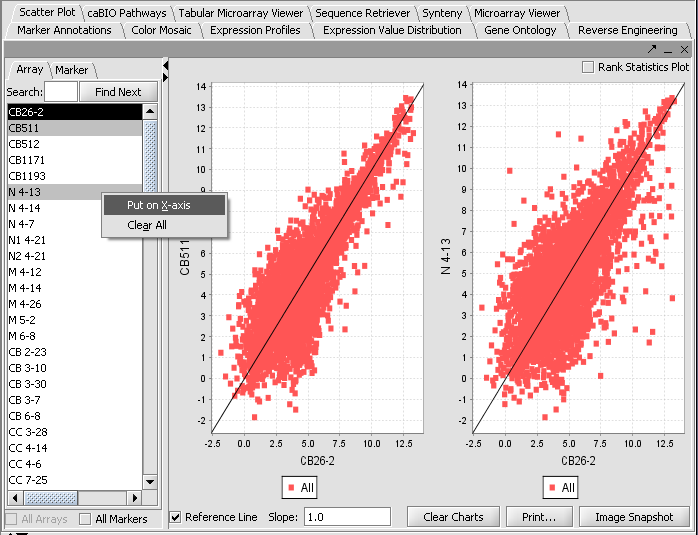
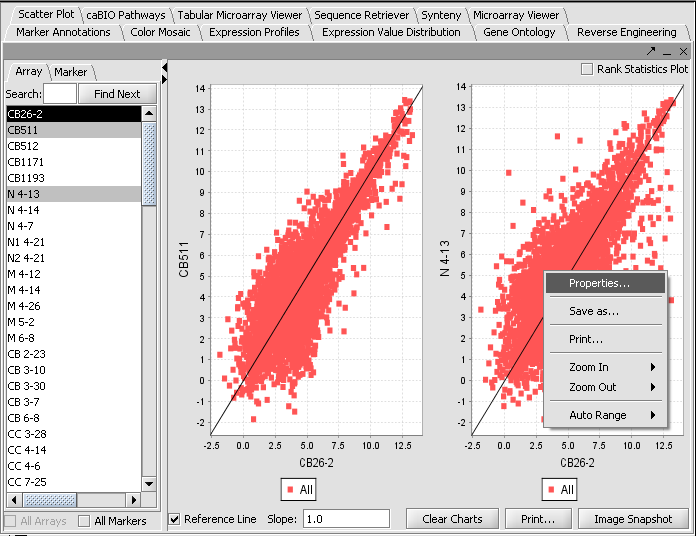
Visualization tools provide a view of the chip(s) under investigation and can be used for ascertaining the quality of the data. Activating gene and phenotype sets (Tutorial - Data Subsets) can be used to restrict the number of markers/arrays displayed. The images created can be saved and exported. A detailed description on how to manipulate visualization components is described in the online help.
Preparation
Load a microarray dataset. This could be the dataset created in Tutorial - Projects and Data Files, or the file webmatrix.exp that is included with the geWorkbench download.
View a dataset
- Select the Microarray Viewer visualization component in the View Area at the top-right section of the interface.
- The All Markers and All Arrays checkboxes can be used to override the effects of activated sets, should they be in use. When no sets are activated, all markers or arrays will be used.
Preferences
The Preferences selection in the Tools menu allows users to specify how certain aspects of the system will behave. Once your preferences are set, these preferences are persistent between application sessions.
Modifying Settings
1. From the main menu, click on Tools>Preferences.
2. In the Preferences pop-up window, you can define settings for:
- Text Editor: The editor selected will be used to open and inspect data sets loaded in a project. Notepad is the default setting.
- Visualization: The color scheme to be applied to color mosaic images.
- Absolute: (default) Let M = max{|min|, |max|} over all expression measurements, across all arrays. If expression value x > 0, assign it the red spectrum x / M * 256. If expression value x is negative, assign it to the green spectrum -x / M * 256.
- Relative: This is similar to the setting for Absolute, but each marker is mean-variance normalized first.
- Genepix Value Computation: You can specify how compute the value displayed for Genepix array. The default setting is Option (Mean F635 - Mean B635) / (Mean F532 - Mean B532).
Select Relative for the visualization preference. Note that this choice will not take effect until the next time you load a data set.
3. Click on OK.