Difference between revisions of "Pattern Discovery"
(→Example Pattern Discovery run) |
(→Setup (BLOSUM50)) |
||
| Line 198: | Line 198: | ||
We can change the Advanced Parameters to use the BLOSUM50 similarity matrix rather than requiring exact matches. | We can change the Advanced Parameters to use the BLOSUM50 similarity matrix rather than requiring exact matches. | ||
| + | |||
| + | To use a substitution matrix, uncheck the "Exact only" box. | ||
[[Image:T_PatternDiscovery_Params_Advanced_BLOSUM.png]] | [[Image:T_PatternDiscovery_Params_Advanced_BLOSUM.png]] | ||
| − | |||
===Result (BLOSUM50) in line view=== | ===Result (BLOSUM50) in line view=== | ||
Revision as of 12:17, 12 February 2010
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Outline
In this tutorial, running a pattern discovery algorithm on a set of protein sequences is described. The steps include:
- Setting parameters
- Creating a new session
- Running the job
- Viewing the results
Overview
The geWorkbench Pattern Discovery module uses an algorithm called SPLASH (Califano, 2000) to search for common patterns in sets of protein or DNA sequences. This type of search could be used, for example, to search for common structural or regulatory elements in otherwise unrelated sequences.
(Note - there currently is no provision for filtering out repeated sequences from genomic seqeuence. Results on DNA sequences should be evaluated in this light).
Component layout
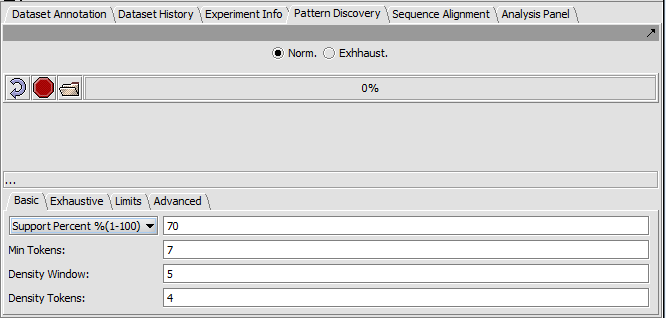
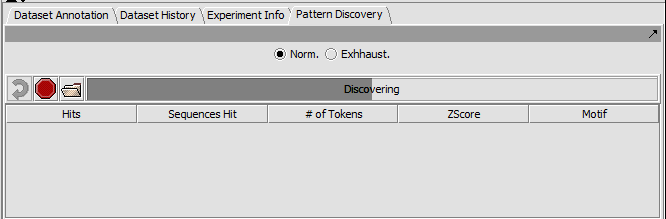
The Pattern discovery component control area has three sections.
- Run controls are shown at top,
- the hit display area is in the center, and
- the parameter settings are at bottom.
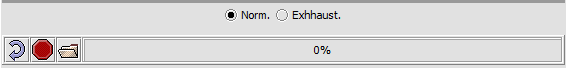
Top-level run controls
- Norm - select the "normal" pattern discovery algorithm.
- Exhaustive - select the "exhaustive" pattern discovery algorithm. This takes much more time.
- Curling arrow - start a new pattern discovery run.
- Stop sign - stop the current pattern discovery run
- File folder - load a pattern file
- Progress bar - To the right of the control buttons is a progress bar that reports on the current stage of discovery being executed.
Setting parameters
A number of parameters can be adjusted by the user to adjust the sensitivity of the search.
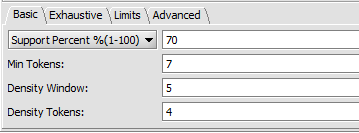
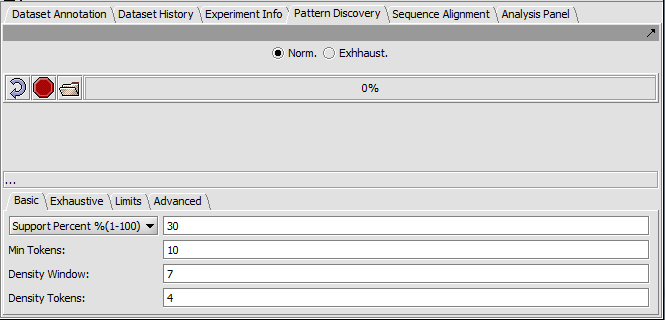
Basic tab
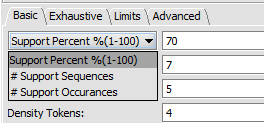
- Support - Can be input in number of sequences or in % of sequences containing a given motif.
- Support Percent % - The pattern must appear in at least given percentage of sequences.
- # Support Sequences - The pattern must appear in at least the given number of sequences.
- # Support Occurances - The pattern must occur at least the given number of times in the set of sequences (can be more than once per sequence).
- Min Tokens - The minimum number of characters in a discovered motif.
- Density Window - A sliding window in which at least the number of tokens set in "Density Tokens" must be found.
- Density Tokens - the minimum number of matching characters within the "Density Window".
Exhaustive tab
Parameters specific to the "exhaustive" search algorithm can be set in this tab.
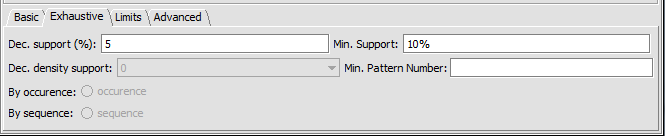
- Dec. support (%) -
- Dec. density support -
- Min support -
- Min pattern number -
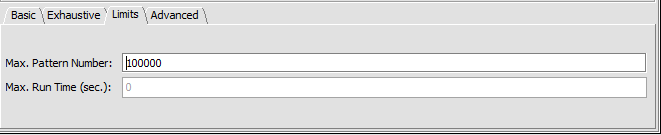
Limits
- Max Pattern Number -
- Max run time (sec) -
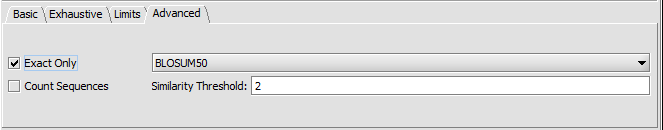
Advanced tab
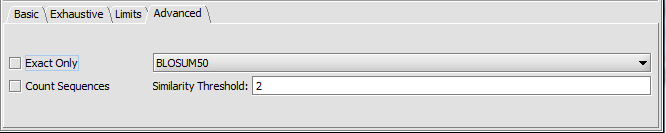
- Exact Only - When checked, no substitution matrix will be used. Exact exact character matches are required. "Exact only" should always be used for DNA as no DNA substition matrix is provided.
- Count Sequences -
- Similarity matrix choice (default BLOSUM50) - Other choices are BLOSUM62 and BLOSUM100.
- Similarity threshold -
Run Pattern Discovery
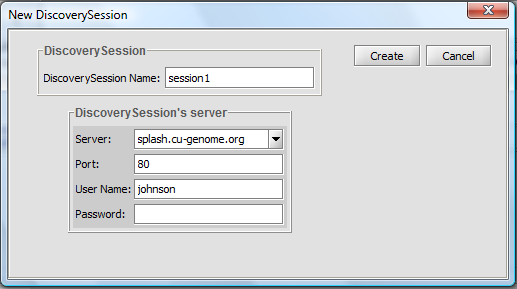
- Pushing on the button with the curling arrow icon will bring up the session creation box:
Discovery Session
- Discovery Session Name: A name is auto-generated for identifying the job on the server, but any name can be entered.
- Create - Push to start the search.
- Cancel- Cancel the discovery run.
Discovery Session Server
- Server and Port: Columbia supports a Pattern Discovery server at splash.cu-genome.org, Port 80.
- Username: Any name can be entered to identify the job.
- Password: none currently required.
The progress bar will show the several stages of a search.
Viewing results
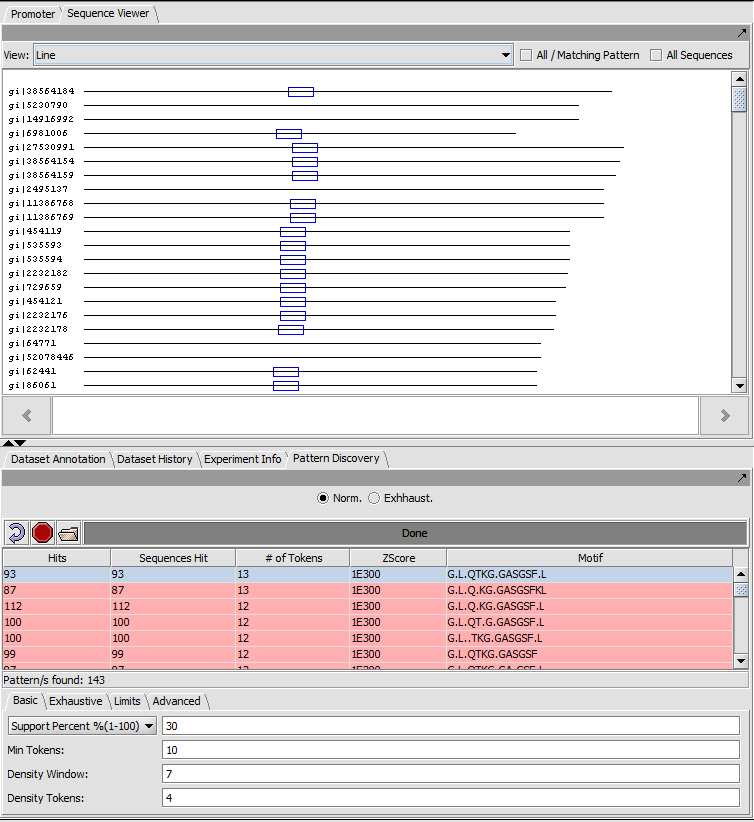
The result of the search can be viewed both in the Pattern Discovery module itself and in other sequence viewer modules such as "Sequence" and "Promoter". In Pattern Discovery the results are returned in a table, and the hits for the motif(s) selected in the table will be displayed superimposed on the sequences above. In this picture, the results were first sorted by the number of tokens.
Adding results to the Projects Folder
The results of a run of Pattern Discovery are automatically placed in the Project Folder:
Options to Save or Mask Patterns
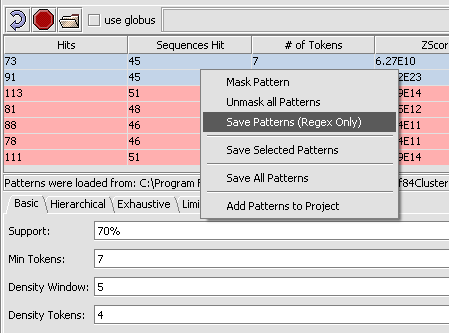
Several operations are possible on the returned patterns. The options menu can be seen by right-clicking on a selection of one or more returned patterns.
1. The patterns can be saved, either with their positions on the original sequences, or just as regular expressions.
2. The patterns can be masked out of the query sequence.
The options shown in the picture below are:
Mask Pattern - The selected pattern(s) will be masked out of the sequence for future searches.
Unmask All Patterns - Undo the masking.
Save Patterns (Regex Only) - This will save the selected pattern(s) in the form of regular expressions, that is, letters and wild-card characters.
Save Selected Patterns - This will save both the selected pattern(s) and their hits to the query sequences. The locations (positions on the query sequences) saved are specific to the particular input file used. The name of this file is saved in the pattern file.
Save All Patterns - This will save both all of the patterns and their hits to the query sequences. The locations positions on the query sequences) saved are specific to the particular input file used. The name of this file is saved in the pattern file.
Add Patterns to Project - This will add the patterns found as a data node in the Projects Folder, with the position information on the input sequences.
Logical complexities in the display...
- The other sequence display components, including Sequence, Promoter, and Position Histogram, are only available when the parent sequence object is selected in the Project Folder. The results of the Pattern Discovery run are still available in that component.
- Selecting another sequence object will cause the Pattern Discovery component to be cleared. However, the results can be reloaded by again selecting the Pattern Discovery result in the Project Folder.
- Pattern Discovery results can only be displayed in the context of the sequences from which they were derived.
References
Califano, A. (2000). SPLASH: structural pattern localization analysis by sequential histograms. Bioinformatics, Apr;16(4):341-57 (link to paper).
Example Pattern Discovery run
Setup (Exact matches)
1. We will use a file containing a number of histone sequences, H1H5_HistoneDB_NHGRI.fasta.
2. We will try parameters set to allow longer matches. No changes from default are made in the other parameter tabs. In particular, this search uses exact matching of the sequence letters, without substitutions.
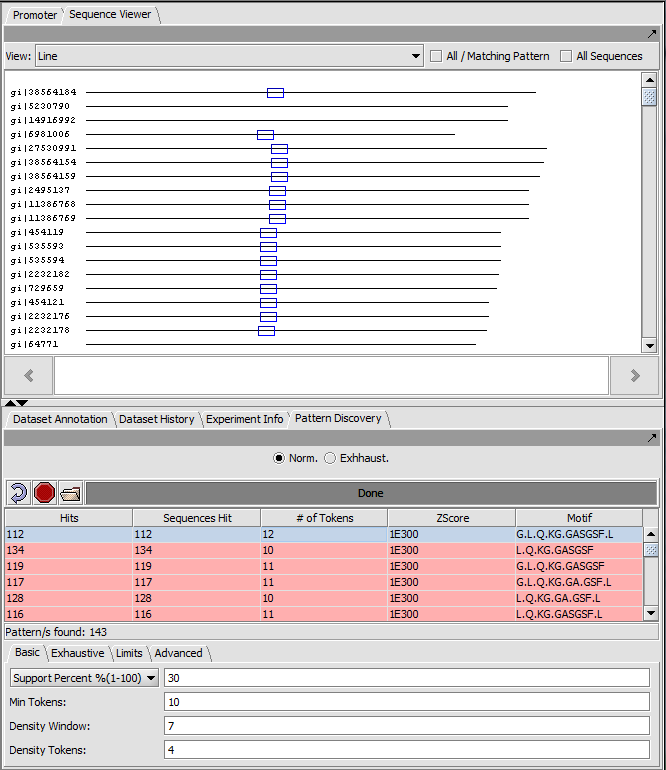
Result (Exact matches) sequence line view
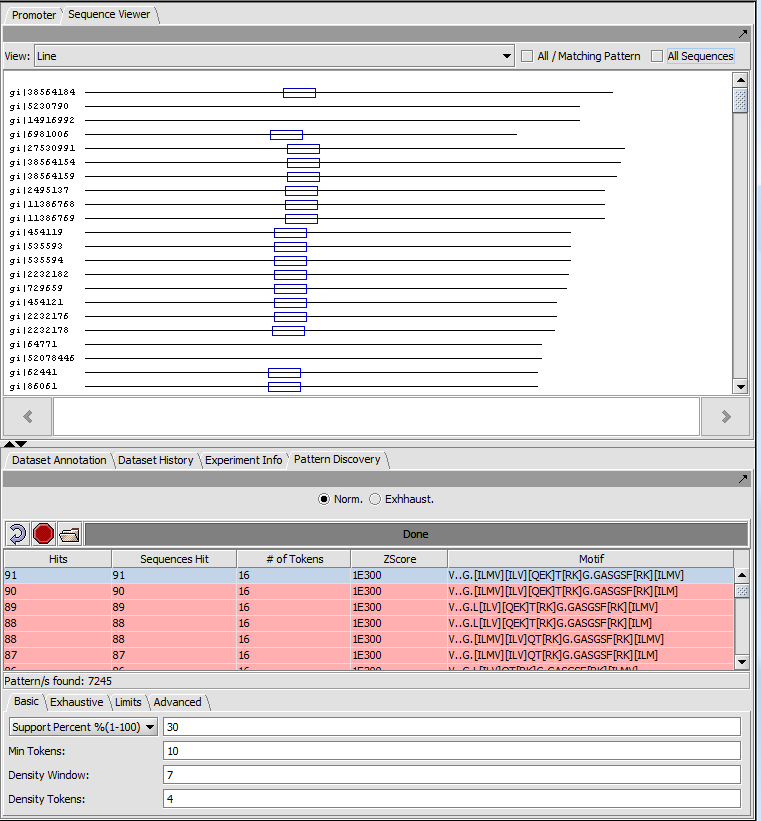
Each sequence is shown as a line with length proportional to the sequence length. All sequences are left-aligned.
The position of matches along the sequence are shown using colored boxes (here blue).
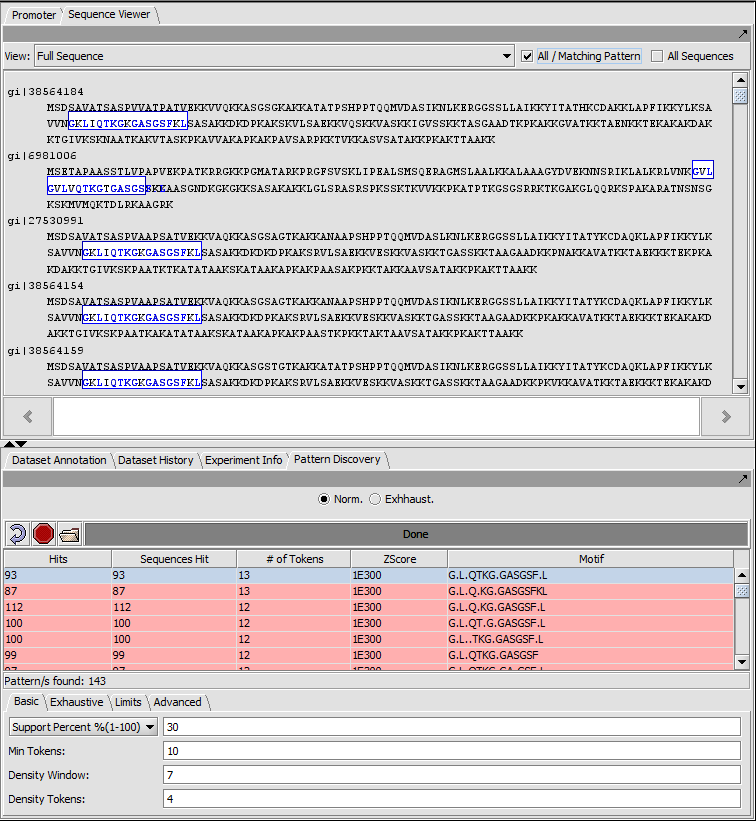
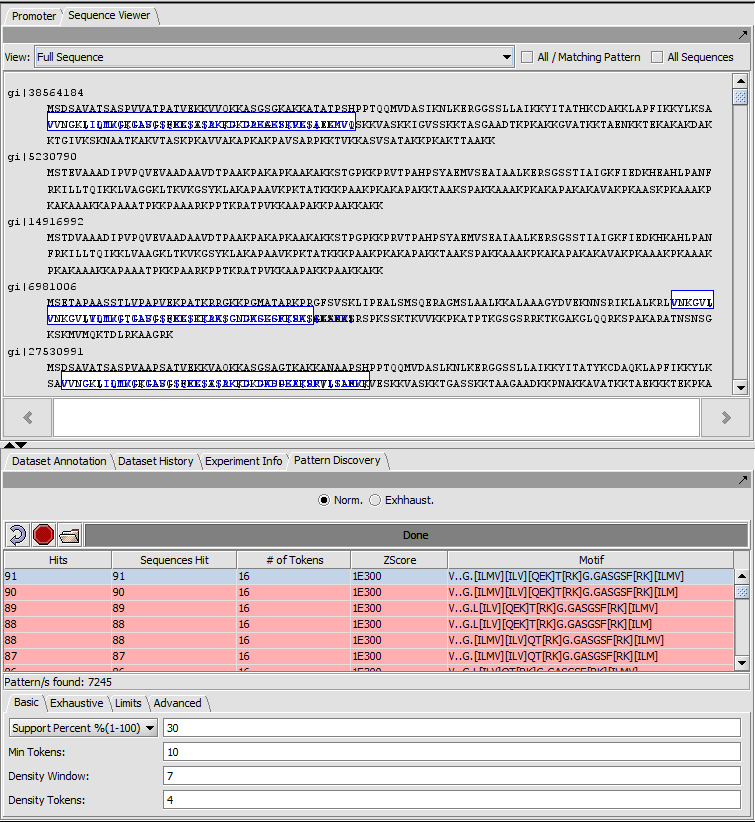
Result (Exact matches) full sequence view
In full sequence view, the actual sequence letters are displayed and matches are again outlined in colored boxes (here blue).
Setup (BLOSUM50)
We can change the Advanced Parameters to use the BLOSUM50 similarity matrix rather than requiring exact matches.
To use a substitution matrix, uncheck the "Exact only" box.