SAM
Contents
SAM - Significance Analysis of Microarrays
Local Installation of R Server
The SAM analysis component can use a local installation of R on your desktop computer.
This has been tested with R version 2.15.0 and 3.0.2.
There are special considerations for installing R on Windows computers, please see R installation on Windows.
Setting the R location in geWorkbench is covered at Preferences: R Location.
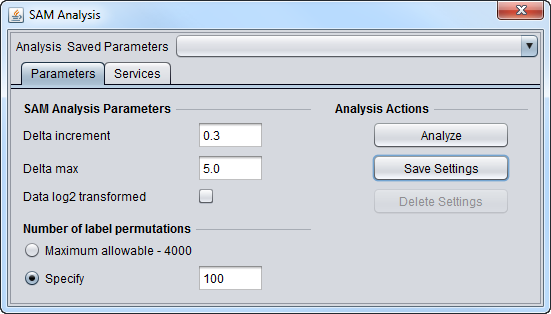
SAM Parameters
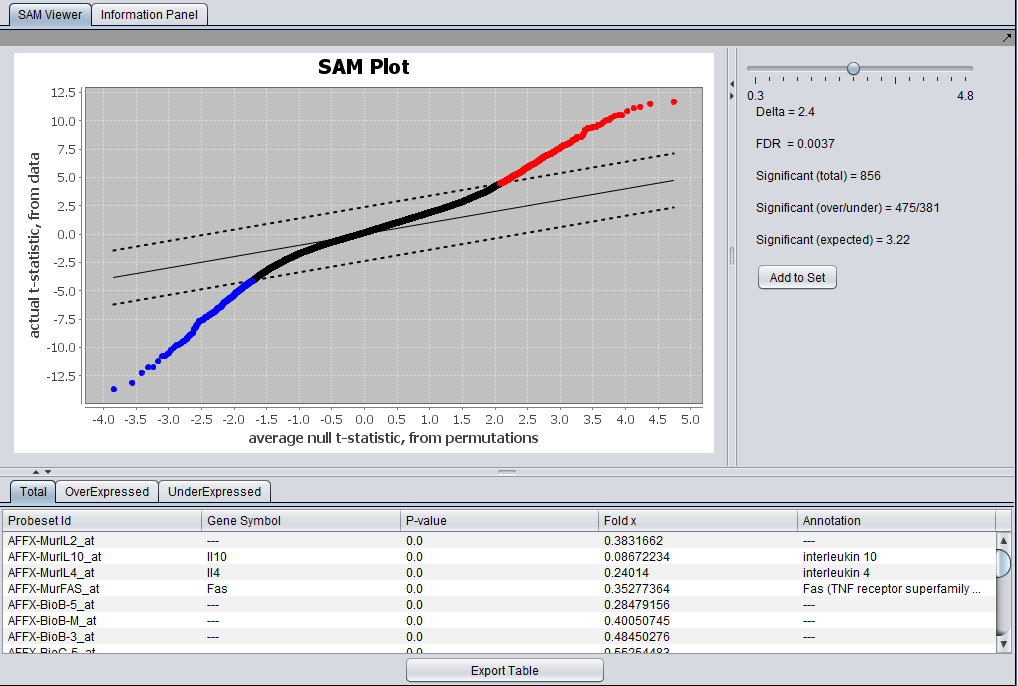
SAM Results Viewer
The Full SAM Reults viewer is shown here.
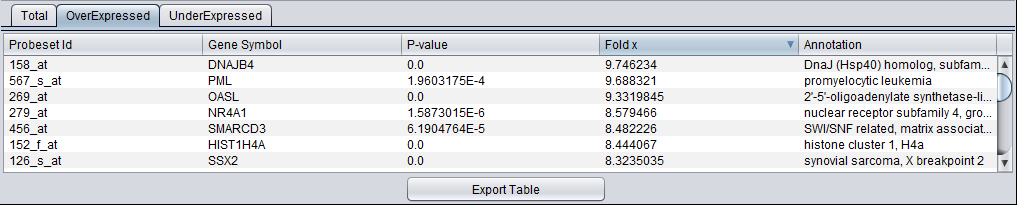
The results can be sorted, e.g. on the fold change value as shown below.