Difference between revisions of "Sequence Retriever"
| Line 20: | Line 20: | ||
Once a set of sequences has been obtained, it can used for several types of analysis in geWorkbench, including searching using known promoter motifs ([[Tutorial_-_Promoter_Analysis | Promoter_Analysis]]), running [[Tutorial_-_BLAST|BLAST]] searches, or looking for common motifs using [[Tutorial_-_Pattern_Discovery|Pattern Discovery]]. | Once a set of sequences has been obtained, it can used for several types of analysis in geWorkbench, including searching using known promoter motifs ([[Tutorial_-_Promoter_Analysis | Promoter_Analysis]]), running [[Tutorial_-_BLAST|BLAST]] searches, or looking for common motifs using [[Tutorial_-_Pattern_Discovery|Pattern Discovery]]. | ||
| − | + | Nucleotide sequences are obtained directly from the UC Santa Cruz Golden Path database. Amino-acid sequences are retrieved from the European Bioinformatics Institute (EBI). | |
| − | |||
| − | |||
| − | |||
| − | |||
==Example - retrieving sequences for a list of gene markers== | ==Example - retrieving sequences for a list of gene markers== | ||
===Obtaining a set of markers=== | ===Obtaining a set of markers=== | ||
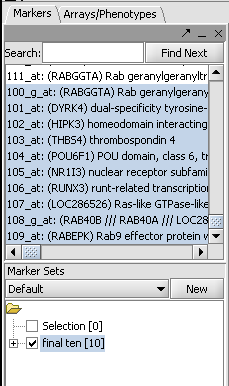
| − | + | Sequences can be retrieved for any set of markers of interest. For this example we have loaded the tutorial data file BCell-100.exp and selected the last 10 markers into a new Marker Set: | |
| − | + | [[Image:T_SequenceRetriever_MarkerSet.png]] | |
| − | |||
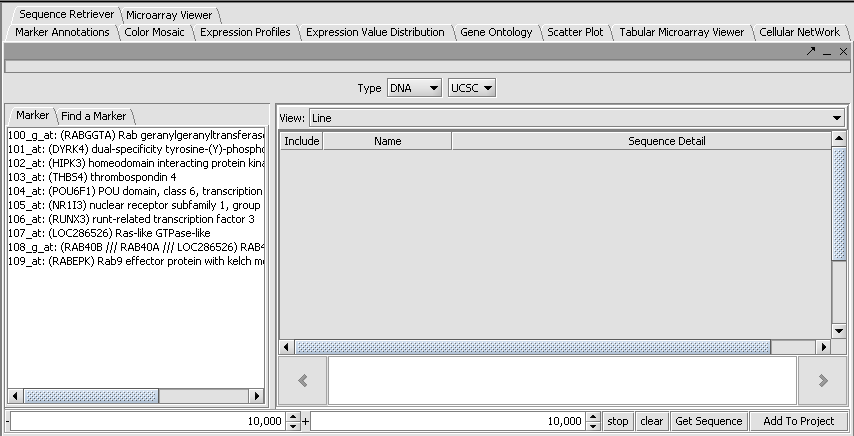
| − | + | When the set is activate (through use of the check box) the selected marker set will appear in the Sequence Retriever component: | |
| + | [[Image:T_SequenceRetriever_Setup.png]] | ||
| − | |||
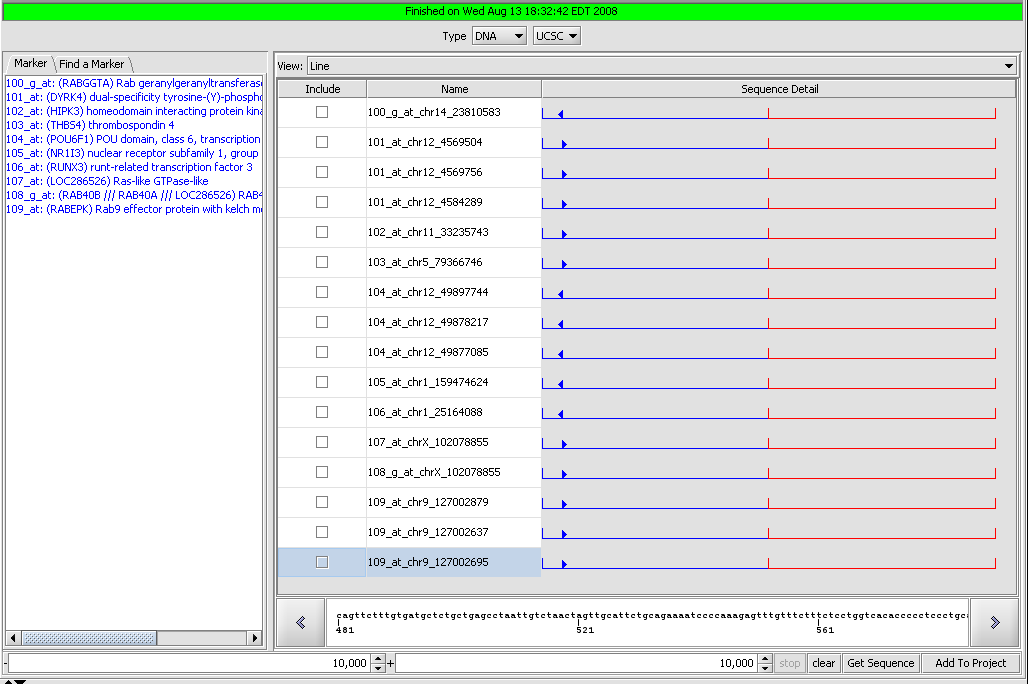
| + | We will retrieve DNA sequences from Santa Cruz and leave the default settings of +-10,000 relative to the start of transcription. After pressing '''Get Sequence''' the sequences are downloaded: | ||
| − | + | [[Image:T_SequenceRetriever_AfterRetrieval.png]] | |
| + | Note that for several of the markers more than one sequence has been retrieved. All sequences associated with a given gene symbol are retrieved. | ||
| − | |||
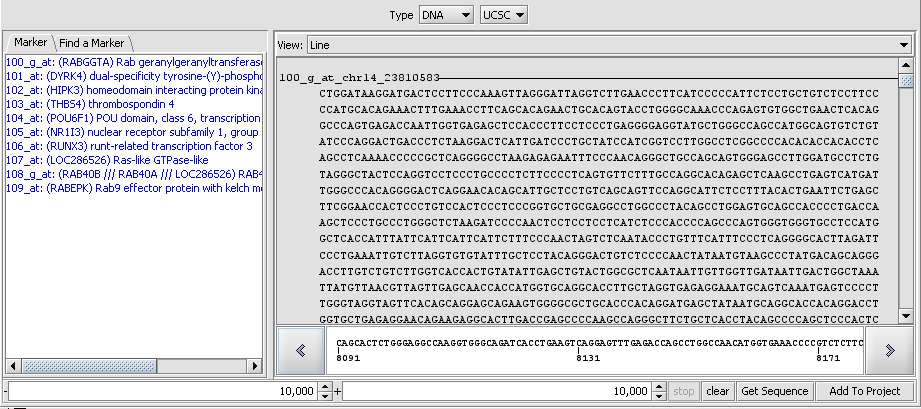
| − | + | Double-clicking on one of the lines shows the sequence detail: | |
| − | + | [[Image:T_SequenceRetriever_SequenceDetail.png]] | |
| − | |||
| − | + | The component provides check boxes which allow sequences of interest to be selected and added to the Project Folders component as a data node: | |
| + | [[Image:T_SequenceRetriever_SelectingForProject.png]] | ||
| − | + | When Add to Project is pushed, the user is asked for a name for the new data node: | |
| − | + | [[Image:T_SequenceRetriever_NamingSet.png]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | The resulting node is placed into the Project Folder as a child of the original dataset: | ||
| − | + | [[Image:T_SequenceRetriever_SequenceNode.png]] | |
| − | |||
| − | |||
| − | |||
| + | Note that when this node is added, the Viewing area of the geWorkbench GUI will now show components that support working with sequences. However, the Sequence Retrieval component will no longer be visible! You must select the Project or the sequence's parent object to see the Sequence Retrieval component again. | ||
| + | ==Saving the sequences to an external FASTA file== | ||
| − | + | # Right-click on the "selected sequences" entry you made in the Project Folders component. | |
| − | + | # Select Save. | |
| − | + | # Enter a suitable name and save the file. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Revision as of 18:09, 13 August 2008
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Outline
In this tutorial, we will
- Discuss uses for retrieved sequences.
- Review obtaining a set of markers for which we wish to retrieve sequences.
- Retrieve DNA sequences from a remote resource
- View the sequences in a sequence browser.
- Add the sequences to the Project Folders component
- Create a new list containing those genes for which sequences were returned.
- Save the list of sequences to a file.
- Save the sequences to a file.
- Note that the different objects in the Project Folders component each have their own sets of Marker Sets.
Overview
geWorkbench contains a number of modules that allow DNA or protein sequences to be visualized and analyzed. Sequences can be loaded from a local disk as a FASTA format file, or can be retrieved from a remote resource. Here we discuss retrieval of sequences from the network.
Once a set of sequences has been obtained, it can used for several types of analysis in geWorkbench, including searching using known promoter motifs ( Promoter_Analysis), running BLAST searches, or looking for common motifs using Pattern Discovery.
Nucleotide sequences are obtained directly from the UC Santa Cruz Golden Path database. Amino-acid sequences are retrieved from the European Bioinformatics Institute (EBI).
Example - retrieving sequences for a list of gene markers
Obtaining a set of markers
Sequences can be retrieved for any set of markers of interest. For this example we have loaded the tutorial data file BCell-100.exp and selected the last 10 markers into a new Marker Set:
When the set is activate (through use of the check box) the selected marker set will appear in the Sequence Retriever component:
We will retrieve DNA sequences from Santa Cruz and leave the default settings of +-10,000 relative to the start of transcription. After pressing Get Sequence the sequences are downloaded:
Note that for several of the markers more than one sequence has been retrieved. All sequences associated with a given gene symbol are retrieved.
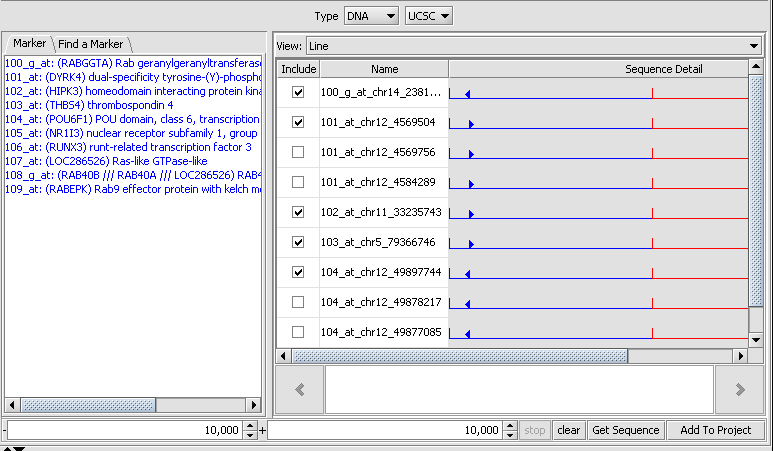
Double-clicking on one of the lines shows the sequence detail:
The component provides check boxes which allow sequences of interest to be selected and added to the Project Folders component as a data node:
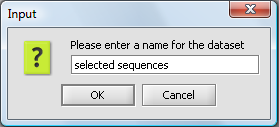
When Add to Project is pushed, the user is asked for a name for the new data node:
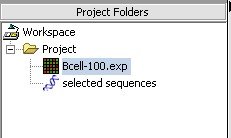
The resulting node is placed into the Project Folder as a child of the original dataset:
Note that when this node is added, the Viewing area of the geWorkbench GUI will now show components that support working with sequences. However, the Sequence Retrieval component will no longer be visible! You must select the Project or the sequence's parent object to see the Sequence Retrieval component again.
Saving the sequences to an external FASTA file
- Right-click on the "selected sequences" entry you made in the Project Folders component.
- Select Save.
- Enter a suitable name and save the file.