Difference between revisions of "Sequence Retriever"
(→Outline) |
(→Overview) |
||
| Line 4: | Line 4: | ||
==Overview== | ==Overview== | ||
| − | + | The Sequence Retriever component fetches the DNA or protein sequences for selected markers from remote databases. Nucleotide sequences are retrieved from the GoldenPath database hosted at the UC Santa Cruz. Amino-acid sequences are retrieved from the European Bioinformatics Institute (EBI). | |
| − | + | One the desired sequences have been downloaded, they can be viewed or saved to the Project Folders component for use by other components, for example [[BLAST|BLAST]], searching using known promoter motifs ([[Promoter_Analysis | Promoter_Analysis]]), or looking for patterns in a set of sequences using [[Pattern_Discovery|Pattern Discovery]]. | |
| − | |||
| − | |||
==Controls== | ==Controls== | ||
Revision as of 10:00, 9 June 2011
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
The Sequence Retriever component fetches the DNA or protein sequences for selected markers from remote databases. Nucleotide sequences are retrieved from the GoldenPath database hosted at the UC Santa Cruz. Amino-acid sequences are retrieved from the European Bioinformatics Institute (EBI).
One the desired sequences have been downloaded, they can be viewed or saved to the Project Folders component for use by other components, for example BLAST, searching using known promoter motifs ( Promoter_Analysis), or looking for patterns in a set of sequences using Pattern Discovery.
Controls
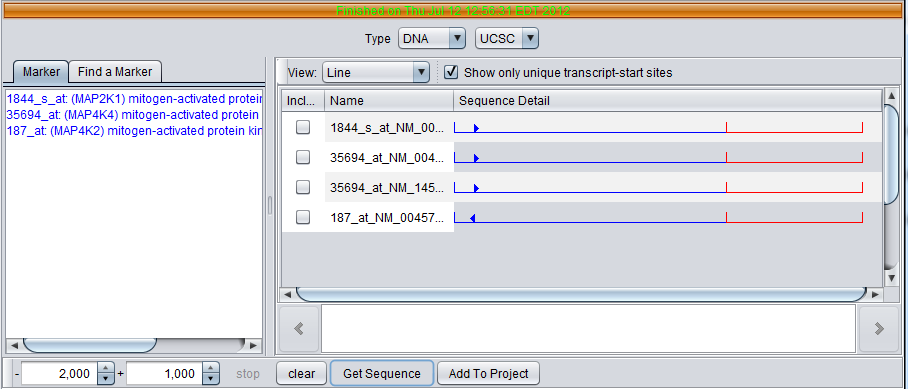
- Type - DNA or Protein
- Source - The pulldown menu to the right of the "Type" pulldown shows the source to query for the data. At present, only one choice for each type is supported. DNA sequences are retrieved from the UCSC Santa Cruz. Protein sequences are retrieve from the EBI.
- Marker - This panel shows markers that are in any activated set in the Markers component.
- Find a Marker - This tab allows one to search in the component for a particular marker. However, we recommend performing searches in the Markers component and adding the results to a set.
- "- and +" text fields - For DNA sequence retrieval, these two text fields specify the distance upstream (-) and downstream (+) from the transcription start site for the request. They are disabled when a protein query is selected. The default setting is 2000 bp upstream and 1000 bp downstream.
- Clear - clear the query results display.
- Get Sequence - Launches the query using the selected markers.
- Add to Project - Add selected sequences returned by the query to the Project Folders component.
- View - Line or Full Sequence.
- Include - Check boxes for sequences to copy to the Project Folders component when "Add to Project" is pushed.
- Name - The name for a sequence is formed by concatenating the marker name with the chromosome number of physical location of the sequence on the chromosome, e.g. in 35694_at_chr2_102314487, "35694_at" is the probeset id, and the sequence is located on chromosome 2 at position 102314487.
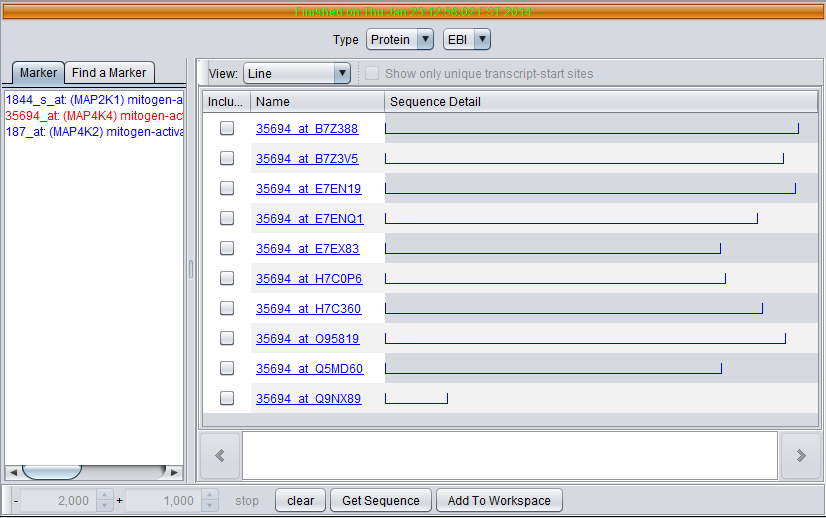
- Sequence Detail - graphically depicts the upstream (blue)and downstream (red) sequences areas, and the transcription start site and direction of transcription (blue arrow).
- Detailed sequence display box - At the bottom of the component, clicking on a sequence in the display above will show the letters with a position scale.
Prerequisites
- A microarray dataset must be loaded.
- An annotation file must be associated with the microarray dataset at the time it is loaded. At present, only Affymetrix-format annotation files can be read in. These files can be obtained for Affymetrix chip types from affymetrix.com. For exact instructions, please see the geWorkbench FAQ page: FAQ
Example - retrieving sequences for a list of gene markers
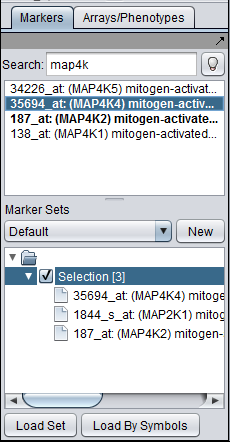
Obtaining a marker to query
Sequences can be retrieved for any set of markers of interest. Here we show searching for MAP4K4 in the Markers component and adding it to the default "Selection" set. When this set is activated, the marker will appear in the Sequence Retriever component.
DNA Query
We will retrieve DNA sequences from Santa Cruz and leave the default settings of -2000 and +1000 relative to the start of transcription.
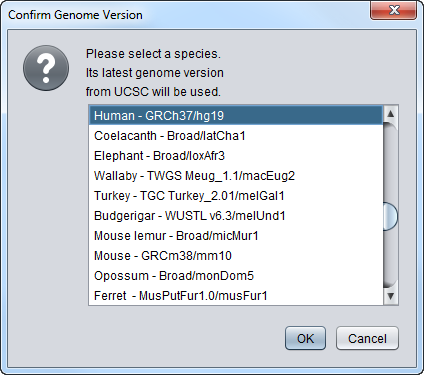
After pressing Get Sequence, if the query is for DNA, the user is prompted to select the genome species and build to query. You should choose the species corresponding to the microarray chip of the dataset you have loaded.
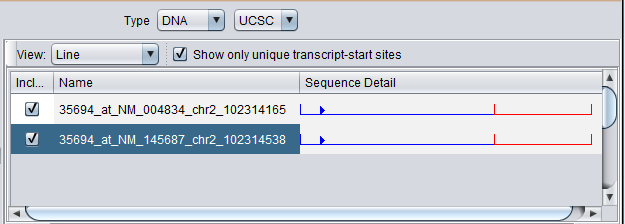
After selecting the appropriate genome, the query will be run and the results displayed
Four genomic sequence were retrieved. All sequences associated with a given gene symbol are retrieved. Each sequence is given a name comprised of the probeset name for the marker, the chromosome, and the location on the chromosome.
The component provides check boxes which allow sequences of interest to be selected and added to the Project Folders component as a data node:
When Add to Project is pushed, the user is asked for a name for the new data node. The resulting node is placed into the Project Folder as a child of the original dataset:
Note that when this node is added, the Viewing area of the geWorkbench GUI will now show components that support working with sequences. However, the Sequence Retrieval component will no longer be visible! You must select the Project or the sequence's parent object to see the Sequence Retrieval component again.
Protein Query
Using the same MAP4K4 marker, query for the protein sequence from EBI.
Six sequences are returned. Note the differences in length as shown graphically.
For protein results, the sequence name is formed by concatenating the probeset id with the UniProt ID of the sequence.
Saving the sequences to an external FASTA file
- Right-click on the "selected sequences" entry you made in the Project Folders component.
- Select Save.
- Enter a suitable name and save the file.