SkyBase
Overview
SkyBase is a database that stores the homology models built by SkyLine analysis for all NESG PSI2 protein structures. Users can search the database with their sequence of interest to find homology models which meet user-defined alignment coverage and sequence identity constraints.
SkyBase Tutorial
http://skybase.c2b2.columbia.edu/nesg3/help/help.html
Database Search Page
http://skybase.c2b2.columbia.edu/nesg3/nesg.php
SkyBase in geWorkbench
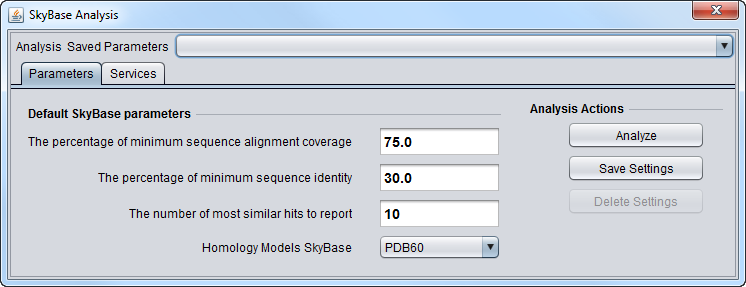
Parameters
- % minimum Alignment Coverage -
- % minimum sequence identity -
- most similar hits to report -
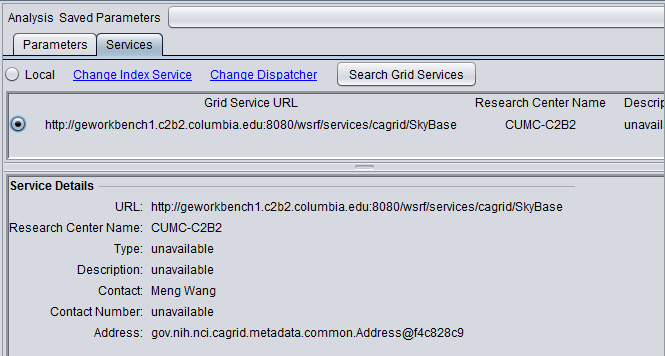
Grid Service
No local service implementation of SkyBase is available in geWorkbench. Instead, an open grid service is used. No username or password is required.
In the Services tab,
- Click on "Search Grid Services". This will retrieve the information for the SkyBase grid service from the index service.
- Select the radio button in front of the SkyBase grid service.
- Return to the "Parameters" tab.
Running a SkyBase query
- Make sure SkyBase is loaded in the Component Configuration Manager.
- Load a protein sequence file for which you wish to find homology models.
- Select the SkyBase analysis component in the Control area of geWorkbench.
- Set the parameters as desired.
- Select the grid service in the "Services" tab.
- Back on the Parameters tab, hit "Analyze".
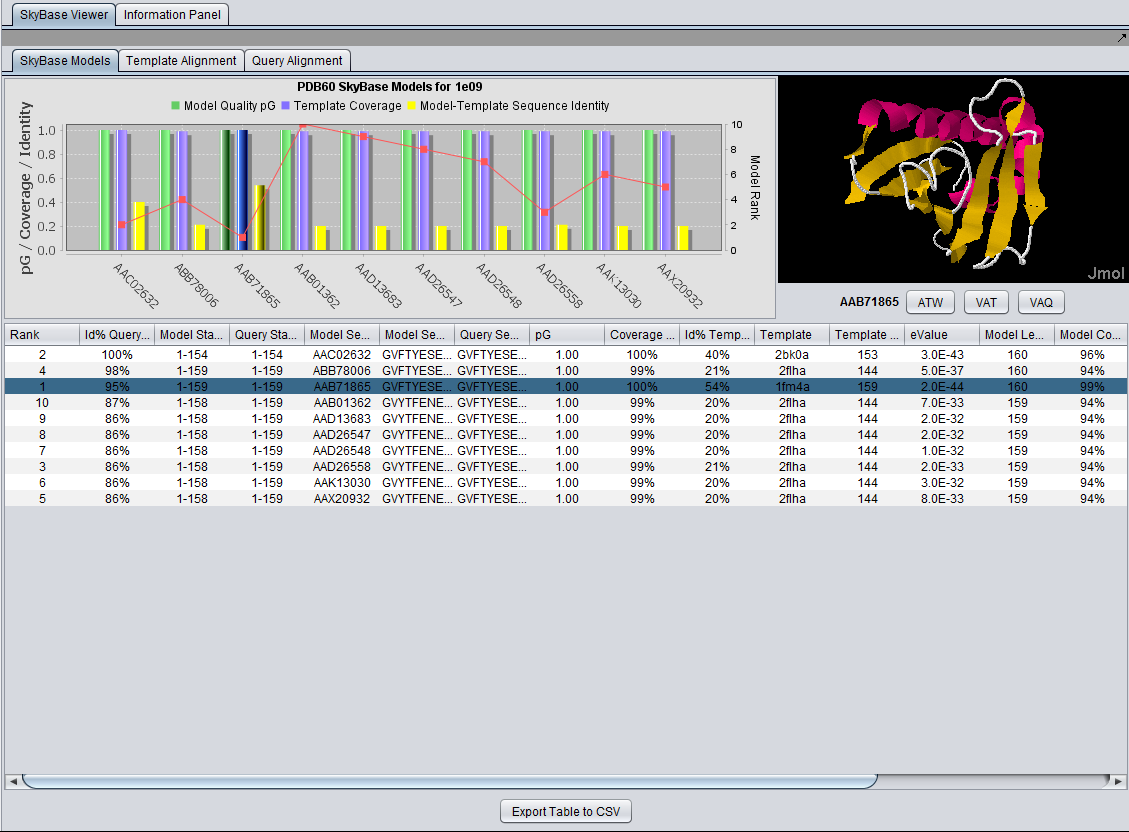
Viewing SkyBase Results
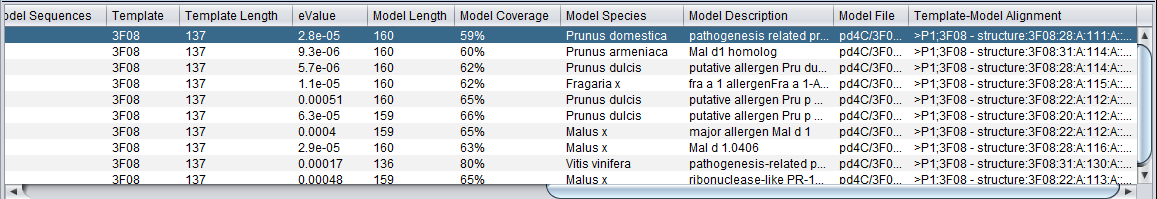
After query with the sequence for PDB structure "1e09", 1e09.fasta:
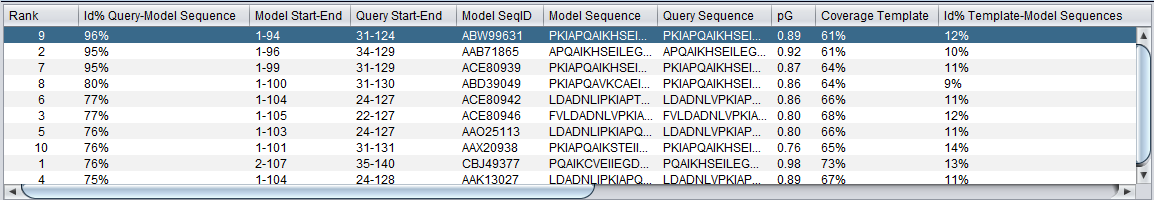
Table
Note on column sorting
In the initial display, the data is sorted in descending order on the second column, "Id% query-model-sequence". The table can be resorted based on any column by clicking on that column's header. Repeated clicks on the same header will cycle through sorting the table in three ways:
- Original order (column 2, descending).
- Ascending order of clicked-on column.
- Descending order of clicked-on column.
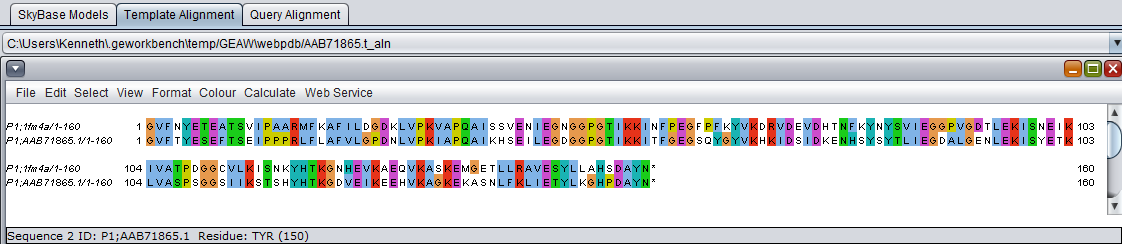
Table Column Details, upper left
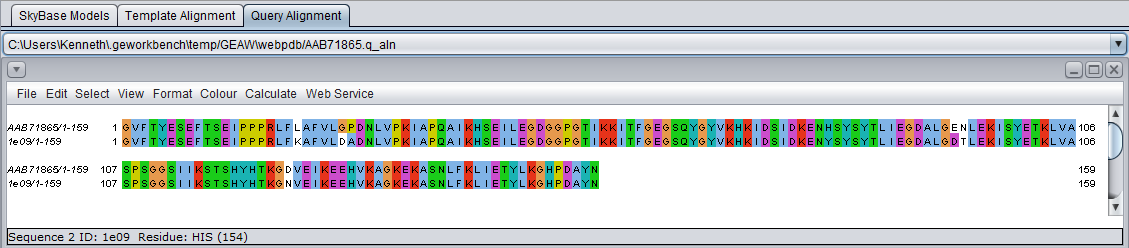
Table Column Details, upper right
VAT
VAQ
References
Lee H, Li Z, Silkov A, Fischer M, Petrey D, Honig B, Murray D. (2010) High-throughput computational structure-based characterization of protein families: START domains and implications for structural genomics. J Struct Funct Genomics. 11(1):51-9. Link to paper
Mirkovic N., Li Z., Parnassa A., Murray D. (2007) Strategies for High-Throughput Comparative Modeling: Applications to Leverage Analysis in Structural Genomics and Protein Family Organization. Proteins: Structure, Function, and Bioinformatics 66:766-777.