Difference between revisions of "Tutorial - Filtering and Normalizing"
(→Filter) |
(→Perform the following steps to filter out data called absent in an Affymetrix file:) |
||
| Line 49: | Line 49: | ||
| − | === | + | ===Filtering out data called absent in an Affymetrix file=== |
| − | + | * Here the file webmatrix.exp has been loaded. It is available on the [[Downloads]] page. It contains unfiltered, unnormalized data. The first microarray dataset, seen in the '''Microarray Viewer''' using the '''Absolute''' display setting, looks like this: | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | [[Image:T_Filtering_Before_Affy_85p.png]] | |
| − | + | ||
| − | + | ||
| − | + | * In the Filtering Panel, select'' Affy Detection Call Filter''. | |
| − | + | * Select ‘A’ (Absent) checkbox and press '''Filter.''' Values that were called '''Absent''' in the original dataset are highlighted in yellow. Internally, the values are now marked as '''Missing'''. | |
| − | + | ||
| − | + | ||
| − | + | [[Image:T_Filtering_After_Affy_85p.png]] | |
| + | |||
| + | |||
| + | For many types of analysis, missing values must be handled properly. The missing values filter allows markers that have more than a specified number of missing values to be removed. Here we will filter out markers for which any values have been marked missing. This would probably be too strict a criterion in many analyses. Another option is to use the '''Missing Value Computation''' found in '''Normalizers''', which will replace missing values with imputed values. | ||
| + | |||
| + | * In the Filtering Panel, select '''Missing Values Filter'''. | ||
| + | * Choose the maximum number of arrays that can have missing values before marker is removed – default is 0. | ||
| + | * Click '''Filter'''. Markers with more than 0 missing values are removed. Notice in the below picture that the yellow values are now gone. | ||
| + | |||
| + | |||
| + | [[Image:T_Filtering_After_MV_85p.png]] | ||
==Normalize== | ==Normalize== | ||
Revision as of 12:52, 8 June 2006
Contents
Filter and Normalize Data
In this tutorial, you will:
- Get acquainted with the various filters and normalizers available in geWorkbench
- Apply a filter and normalizer on a tutorial dataset
Before you can continue, geWorkbench should be running. Load a microarray data file such as webmatrix.exp or cardiomyopathy.exp . Please refer to Tutorial - Projects and Data Files tutorial if you need assistance loading a file.
Filter
Filtering can be used to remove low quality data or reduce the size of the dataset by removing less interesting data. Available geWorkbench filters are as follows:
| Filter | Description | |
|---|---|---|
| Affy Detection Call | Applicable to Affymetrix data only. Sets all measurements whose detection status is any user-defined combination of A, P or M as missing. | |
| Missing values | Discards all markers that have “missing” measurements in at least n microarrays, where n is defined by the user. Another filter must first be applied however, in order to generate the missing values upon which this filter can operate. | |
| Deviation | Marks as missing all markers whose measurements deviate below a given value across all microarrays. | |
| Expression Threshold | Marks as missing all markers whose measurements are inside (or outside) a user-defined range. | |
| 2 Channel | Applicable to 2-channel arrays (Genepix) data only. Defines applicable ranges for each channel, and marks as missing all expression measurements for which either channel intensity is inside (or outside) the defined range. |
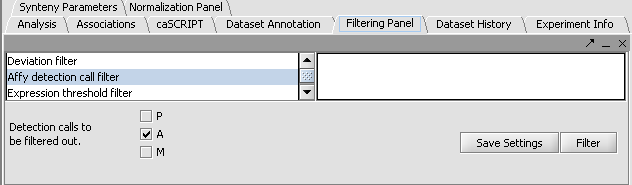
Filtering out data called absent in an Affymetrix file
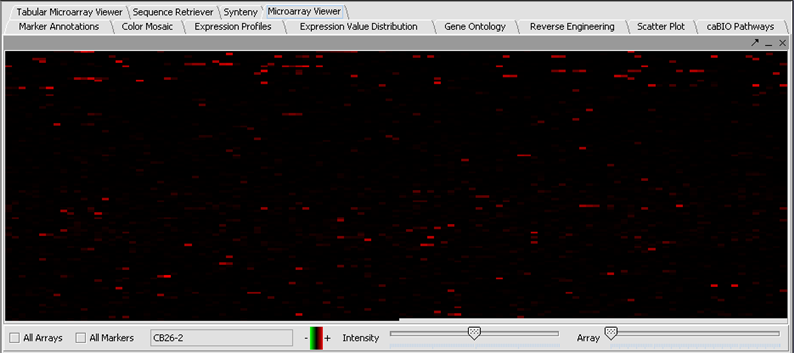
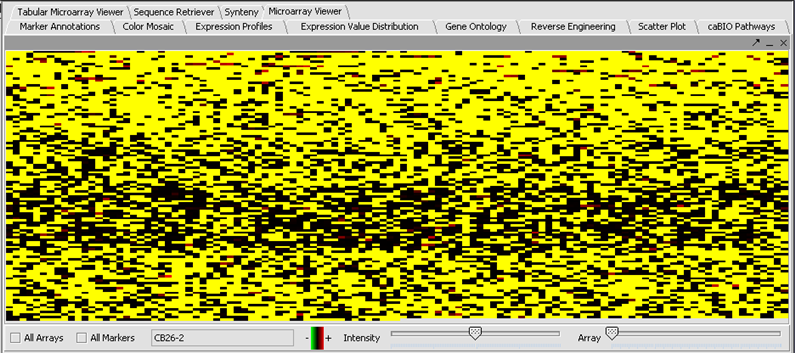
- Here the file webmatrix.exp has been loaded. It is available on the Downloads page. It contains unfiltered, unnormalized data. The first microarray dataset, seen in the Microarray Viewer using the Absolute display setting, looks like this:
- In the Filtering Panel, select Affy Detection Call Filter.
- Select ‘A’ (Absent) checkbox and press Filter. Values that were called Absent in the original dataset are highlighted in yellow. Internally, the values are now marked as Missing.
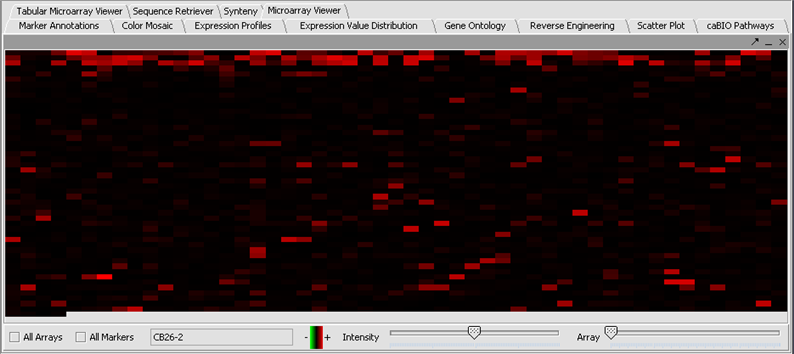
For many types of analysis, missing values must be handled properly. The missing values filter allows markers that have more than a specified number of missing values to be removed. Here we will filter out markers for which any values have been marked missing. This would probably be too strict a criterion in many analyses. Another option is to use the Missing Value Computation found in Normalizers, which will replace missing values with imputed values.
- In the Filtering Panel, select Missing Values Filter.
- Choose the maximum number of arrays that can have missing values before marker is removed – default is 0.
- Click Filter. Markers with more than 0 missing values are removed. Notice in the below picture that the yellow values are now gone.
Normalize
Normalization can be used to decrease the effects of systematic differences across a set of experiments. In geWorkbench, normalization results in replacing values with new values. Available geWorkbench normalization methods are as follows:
| Normalizer | Description | |
|---|---|---|
| Missing value calculation | Replaces every missing value with either the mean value of that marker across all microarrays or with the mean measurement of all markers in the microarray where the missing value is observed | |
| Log2 Transformation | Applies a log2 transformation to all measurements in a microarray | |
| Threshold Normalizer | All data points whose value is less than (or greater than) a user-specified minimum (maximum) value are raised (reduced) to that minimum (maximum) value | |
| Marker-based Centering | Subtracts the mean (median) measurement of a marker profile from every measurement in the profile | |
| Array-based centering | Subtracts the mean (median) measurement of a microarray from every measurement in that microarray | |
| Mean-variance normalizer | For every marker profile, the mean measurement of the entire profile is subtracted from each measurement in the profile and the resulting value is divided by the standard deviation |
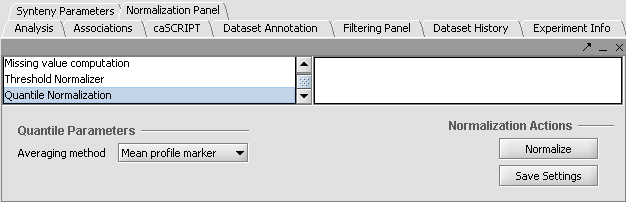
Apply Quantile Normalizer
1. In the Normalization Panel, select Quantile Normalizer.
2. Leave the default averaging method of Mean Profile Marker to indicate handling of missing values..
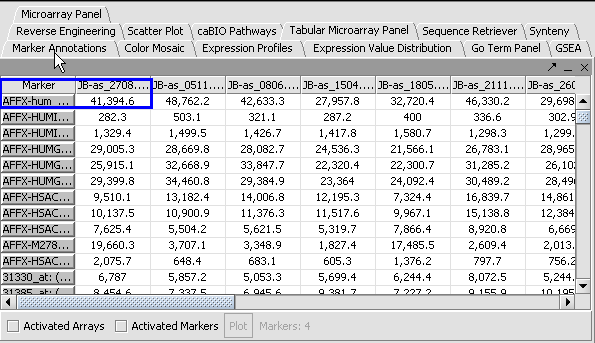
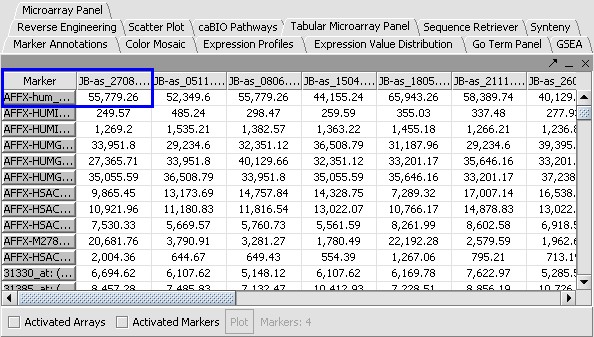
3. Click Normalize. The View Area is updated to reflect normalization (after the screen has been refreshed). Note: The first value in the second row was update from 41,394.6 to 55,779.26.
| PRENORMALIZATION | NORMALIZED | |
|---|---|---|
 |
 . .
|