Tutorial - Network Browser
Contents
Outline
Overview
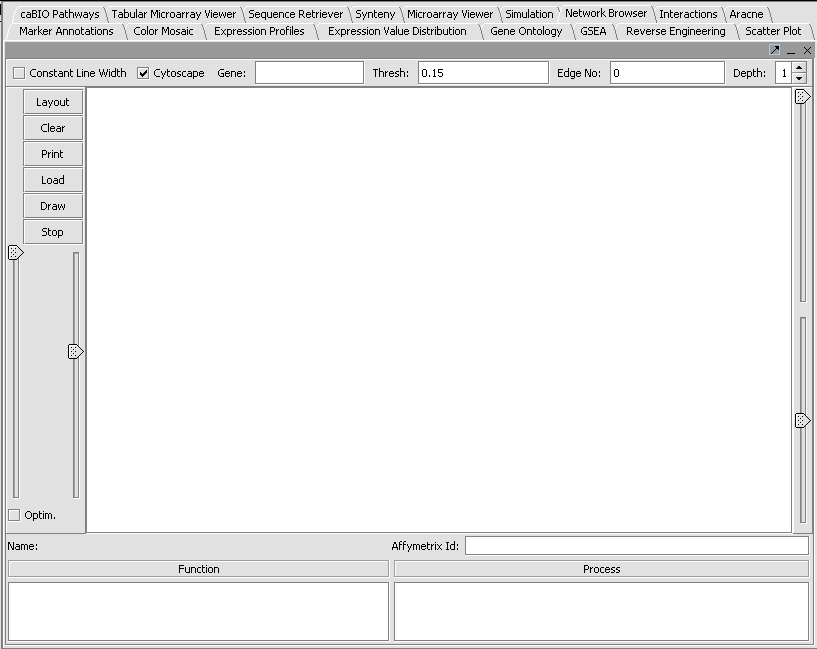
This is a quick introduction to the Network Browser component. This is a beta-release to provide visualizations for adjacency matrices generated by the ARACNE network reverse engineering compoenent and the Interactions component.
Example
- Click the Load button and browse to an adjancency file you would like to load.
- Push the Draw button.
You will see that the component indicates the number of nodes and edges, but no picture is drawn. This is due to an unresolved problem with Cytoscape integration.
Workaround
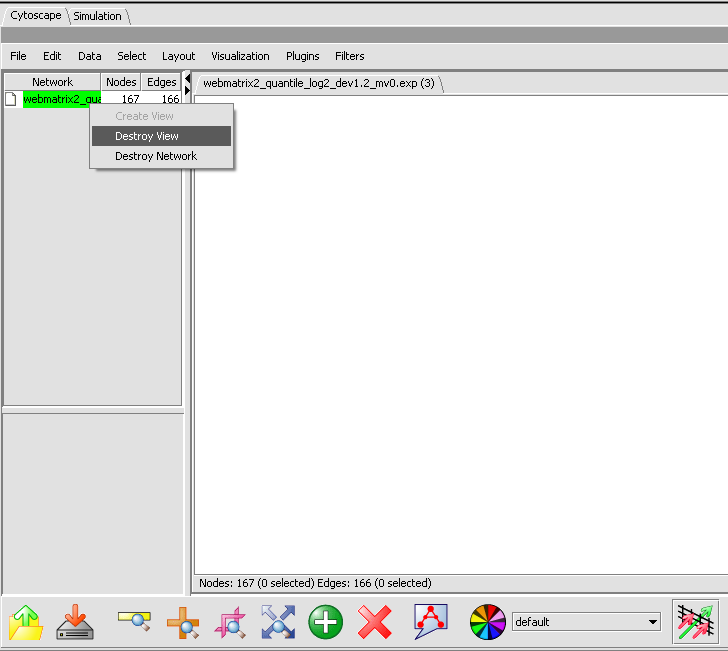
You must destroy and then recreate the view.
- Under the word Network at upper left, right-click on the file name of the adjacency matrix.
- Select Destroy View, as shown below.
- Repeat, and Select Create View.
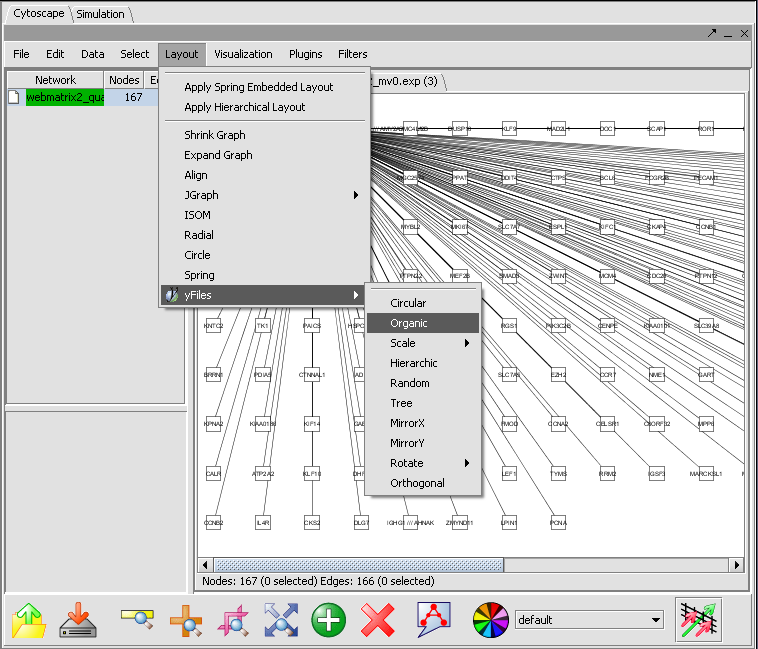
After the network has been displayed, improve the layout by selecting Layout->yFiles->Organic:
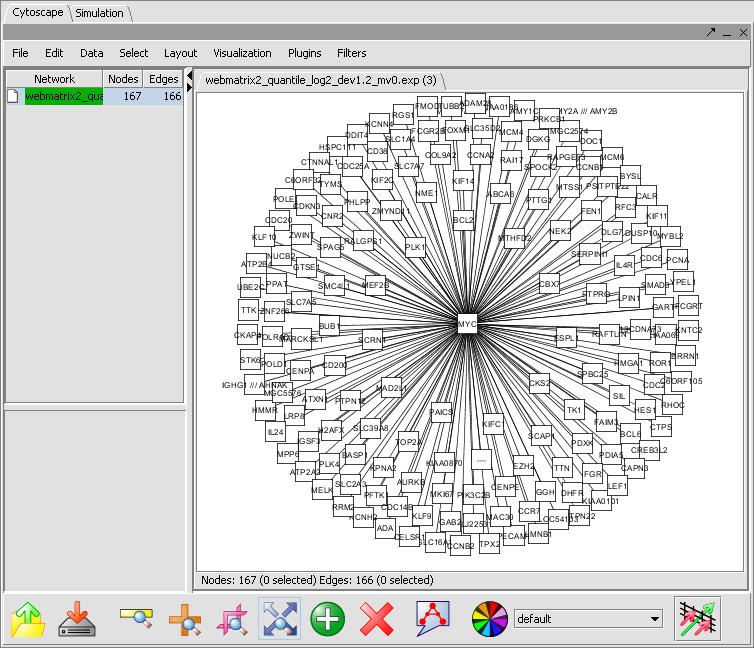
Afther the layout is applied, the network will appear as follows: