Difference between revisions of "Workspace"
(→Overview) |
|||
| (168 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{TutorialsTopNav}} | {{TutorialsTopNav}} | ||
| − | == | + | ==Overview== |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | The Workspace is located in the upper-left-hand corner of the application. It is used to contain open data files and store analysis results during a geWorkbench session. When geWorkbench is launched, an empty Workspace folder is displayed. | ||
| − | + | [[Image:Workspace.png]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | The workspace as a whole, with all its projects and data nodes, can be saved and restored. However, only one workspace can be open at one time. Creating a new workspace or loading a saved workspace will overwrite the current workspace. | ||
| − | |||
| − | |||
| − | |||
| + | * To view the next level down in the hierarchy, click on the “+” icon to expand the branch. | ||
| − | + | * To collapse a branch, click on the “-” icon. | |
| − | |||
| + | The Workspace may contain several heterologous datasets. These datasets can include input (source) data and derived data (results) associated with an experiment as well as image files. Source data can be loaded from the user’s local storage or from remote servers. Loading datasets into the geWorkbench Workspace does not change their physical storage locations. | ||
| − | ''' | + | '''Note''' - The top menu-bar items [[Menu_Bar#File|File]] and [[Menu_Bar#Edit|Edit]] also apply to items in the Workspace. They offer many of the same options shown below, except e.g. Microarray merging is only available from the top level [[Menu_Bar#File|File]] menu. |
| − | + | ==Workspace Menu Options== | |
| + | Right-clicking on the Workspace node gives a menu with the following options | ||
| − | |||
| − | + | [[Image:Workspace_right_click_menu.png]] | |
| − | |||
| − | + | ===Open Files=== | |
| + | * Loading data from local files is covered in the chapter [[Local_Data_Files | Local Data Files]] | ||
| + | * Retrieving data from remote sources (caArray) is covered in [[Remote_Data_Sources | Remote Data Sources]] | ||
| − | |||
| − | + | ===Open PDB File from RCSB Protein Data Bank=== | |
| + | If '''Open PDB File from RCSB Protein Data Bank''' is chosen, a dialog box appears. | ||
| − | |||
| + | [[Image:Project_Folders_Project_Open_RCSB_PDB.png]] | ||
| − | |||
| − | + | Type in the name of a PDB structure entry and it will be retrieved from the RCSB Protein Data Bank and loaded into geWorkbench. | |
| − | |||
| + | ==Workspace Data Node Menu Options== | ||
| + | Right-clicking on a data node will produce a popup menu with the following options: | ||
| − | |||
| − | + | [[Image:Workspace_data_node_right_click.png]] | |
| − | |||
| − | + | ===Save=== | |
| − | + | Save the currently selected data node. This is implemented for at least the below data types. If saving of a particular type has not been implemented, the "Save" option will be disabled (grayed-out). | |
| − | + | * '''Microarray gene expression''' - data is saved using the geWorkbench ".exp" format, regardless of the original format. This allows saving e.g. a merged dataset, and/or any array and marker sets that may have been created. | |
| + | * '''FASTA''' - saved in FASTA format (.fasta). | ||
| + | * '''PDB''' - saved in PDB format (.pdb). | ||
| + | * '''Network''' - saved using the Adjacency Matrix "ADJ" format (.adj). | ||
| + | * '''t-test result''' - saved as comma separated value (.csv) file. | ||
| + | * '''Image''' - saved as PNG file (.png). | ||
| − | + | For each file type, a file browser with a filter for the target file type extension (e.g. .fasta) will be opened. | |
| − | + | ===Export to tab-delim=== | |
| + | This option will only appear for microarray gene expression datasets. It allows the microarray dataset to be exported in a spreadsheet format, as a tab-delimited text file. The first row contains array names and the first column contains the marker names. | ||
| − | + | This export format does not preserve array or marker sets that may have been defined in geWorkbench for the dataset. However, it can be used to save a copy of e.g. merged, filtered, and/or normalized data in a format easily used by other programs. | |
| − | + | When exporting, the file save dialog will display the name of the dataset, minus any recognized file-type suffixes that may be present (e.g. .soft). | |
| − | + | ===Rename=== | |
| − | + | A dialog box will appear in which a new name can be entered. | |
| − | + | [[Image:Workspace_Rename_Node.png]] | |
| + | ===Remove=== | ||
| − | + | The selected data nodes and any child data nodes will be removed. Multiple selections can be made. | |
| − | + | ==Data Node Hover-text Information== | |
| − | + | For microarray datasets, adjacency matrices (network nodes), sequence and pattern nodes, moving the mouse cursor over the data nodes will display additional details about a dataset. | |
| − | + | Microarray datasets: hover text displays number of markers and arrays. | |
| − | |||
| − | + | [[Image:Dataset_hover_microarray.png]] | |
| − | + | Adjacency Matrix: hover text displays number of nodes and edges in the network. | |
| − | |||
| − | + | [[Image:Workspace_Dataset_hover_network.png]] | |
| − | |||
| − | + | Sequence node: | |
| − | |||
| + | [[Image:Workspace_hover_sequences.png]] | ||
| + | Pattern node: | ||
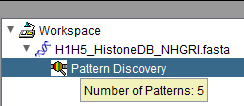
| − | |||
| + | [[Image:Workspace_Pattern_Hover.png]] | ||
| − | + | ==Workspaces== | |
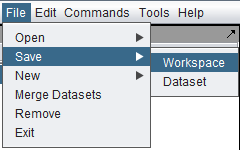
| − | + | ===Saving the Workspace=== | |
| − | + | Saving the workspace saves all its data to a file on disk. The workspace can later be reloaded to resume work. | |
| − | ''' | + | ====Special considerations on saving and restoring workspaces==== |
| + | * '''Versions''' - Workspaces in general may not be compatible across different versions of geWorkbench. | ||
| + | * '''Loaded components''' - The configuration in the [[Component_Configuration_Manager| CCM]] of which components are loaded and which are not is not saved when the workspace is saved; it is maintained separately. | ||
| + | * '''Changes to loaded components''' - If a workspace is saved, and then changes are made to which components are loaded in the [[Component_Configuration_Manager| CCM]], then in rare cases problems may occur when one attempts to reload the saved workspace. | ||
| − | + | [[Image:File_Save_Workspace.png]] | |
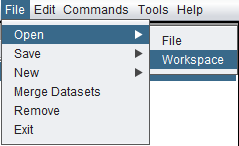
| − | + | ===Opening a Saved Workspace=== | |
| − | + | File->Open-Workspace. | |
| − | + | Only one workspace at a time can be loaded in geWorkbench. Opening a saved workspace will destroy the existing workspace. For this reason, if you opt to open a workspace, you will be prompted as to whether to save the existing workspace first. | |
| − | + | [[Image:File_Open_Workspace.png]] | |
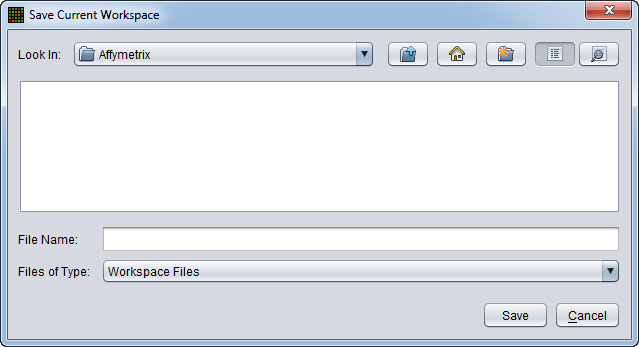
| − | |||
| − | + | A dialog box will appear in which the location and file name to which to save the workspace can be chosen. | |
| − | [[Image: | + | [[Image:File_Save_Workspace_Dialog.png]] |
| − | + | ===Creating a New Workspace=== | |
| − | + | Only one workspace at a time can be loaded in geWorkbench. Creating a new workspace will destroy the existing workspace. For this reason, if you opt to create a new workspace, you will be prompted as to whether to save the existing workspace first. | |
| − | + | A new workspace can only be created from the top level menu bar. | |
| − | |||
| − | + | [[Image:File_New_Workspace.png]] | |
| − | ''' | + | Select '''File->New->Workspace'''. |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Latest revision as of 17:07, 10 January 2014
Contents
Overview
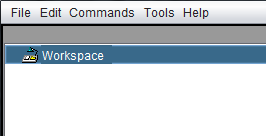
The Workspace is located in the upper-left-hand corner of the application. It is used to contain open data files and store analysis results during a geWorkbench session. When geWorkbench is launched, an empty Workspace folder is displayed.
The workspace as a whole, with all its projects and data nodes, can be saved and restored. However, only one workspace can be open at one time. Creating a new workspace or loading a saved workspace will overwrite the current workspace.
- To view the next level down in the hierarchy, click on the “+” icon to expand the branch.
- To collapse a branch, click on the “-” icon.
The Workspace may contain several heterologous datasets. These datasets can include input (source) data and derived data (results) associated with an experiment as well as image files. Source data can be loaded from the user’s local storage or from remote servers. Loading datasets into the geWorkbench Workspace does not change their physical storage locations.
Note - The top menu-bar items File and Edit also apply to items in the Workspace. They offer many of the same options shown below, except e.g. Microarray merging is only available from the top level File menu.
Workspace Menu Options
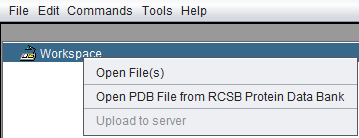
Right-clicking on the Workspace node gives a menu with the following options
Open Files
- Loading data from local files is covered in the chapter Local Data Files
- Retrieving data from remote sources (caArray) is covered in Remote Data Sources
Open PDB File from RCSB Protein Data Bank
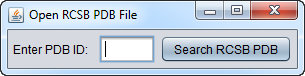
If Open PDB File from RCSB Protein Data Bank is chosen, a dialog box appears.
Type in the name of a PDB structure entry and it will be retrieved from the RCSB Protein Data Bank and loaded into geWorkbench.
Workspace Data Node Menu Options
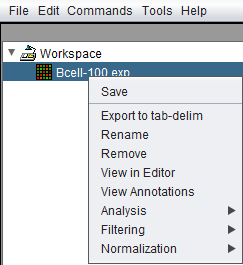
Right-clicking on a data node will produce a popup menu with the following options:
Save
Save the currently selected data node. This is implemented for at least the below data types. If saving of a particular type has not been implemented, the "Save" option will be disabled (grayed-out).
- Microarray gene expression - data is saved using the geWorkbench ".exp" format, regardless of the original format. This allows saving e.g. a merged dataset, and/or any array and marker sets that may have been created.
- FASTA - saved in FASTA format (.fasta).
- PDB - saved in PDB format (.pdb).
- Network - saved using the Adjacency Matrix "ADJ" format (.adj).
- t-test result - saved as comma separated value (.csv) file.
- Image - saved as PNG file (.png).
For each file type, a file browser with a filter for the target file type extension (e.g. .fasta) will be opened.
Export to tab-delim
This option will only appear for microarray gene expression datasets. It allows the microarray dataset to be exported in a spreadsheet format, as a tab-delimited text file. The first row contains array names and the first column contains the marker names.
This export format does not preserve array or marker sets that may have been defined in geWorkbench for the dataset. However, it can be used to save a copy of e.g. merged, filtered, and/or normalized data in a format easily used by other programs.
When exporting, the file save dialog will display the name of the dataset, minus any recognized file-type suffixes that may be present (e.g. .soft).
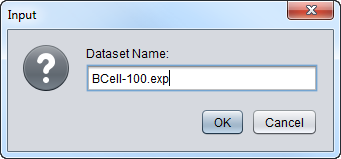
Rename
A dialog box will appear in which a new name can be entered.
Remove
The selected data nodes and any child data nodes will be removed. Multiple selections can be made.
Data Node Hover-text Information
For microarray datasets, adjacency matrices (network nodes), sequence and pattern nodes, moving the mouse cursor over the data nodes will display additional details about a dataset.
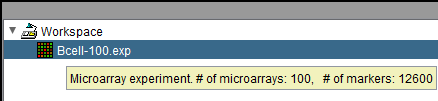
Microarray datasets: hover text displays number of markers and arrays.
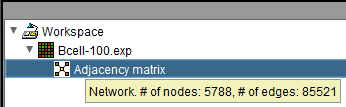
Adjacency Matrix: hover text displays number of nodes and edges in the network.
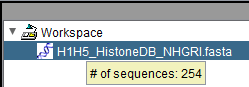
Sequence node:
Pattern node:
Workspaces
Saving the Workspace
Saving the workspace saves all its data to a file on disk. The workspace can later be reloaded to resume work.
Special considerations on saving and restoring workspaces
- Versions - Workspaces in general may not be compatible across different versions of geWorkbench.
- Loaded components - The configuration in the CCM of which components are loaded and which are not is not saved when the workspace is saved; it is maintained separately.
- Changes to loaded components - If a workspace is saved, and then changes are made to which components are loaded in the CCM, then in rare cases problems may occur when one attempts to reload the saved workspace.
Opening a Saved Workspace
File->Open-Workspace.
Only one workspace at a time can be loaded in geWorkbench. Opening a saved workspace will destroy the existing workspace. For this reason, if you opt to open a workspace, you will be prompted as to whether to save the existing workspace first.
A dialog box will appear in which the location and file name to which to save the workspace can be chosen.
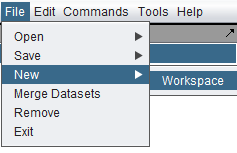
Creating a New Workspace
Only one workspace at a time can be loaded in geWorkbench. Creating a new workspace will destroy the existing workspace. For this reason, if you opt to create a new workspace, you will be prompted as to whether to save the existing workspace first.
A new workspace can only be created from the top level menu bar.
Select File->New->Workspace.