Difference between revisions of "Workspace"
| Line 180: | Line 180: | ||
| − | ===Loading data from a remote instance of caArray | + | ===Loading data from a remote instance of caArray=== |
Click on the Go button next to the caArray data source at the bottom of the dialog. All available caArray experiments will be displayed. | Click on the Go button next to the caArray data source at the bottom of the dialog. All available caArray experiments will be displayed. | ||
| Line 199: | Line 199: | ||
| − | ===To | + | ===To add a remote source=== |
'''1.''' Click on the '''Add A New Resource''' button. | '''1.''' Click on the '''Add A New Resource''' button. | ||
| Line 214: | Line 214: | ||
| − | ===To | + | ===To modify a remote source=== |
The specification of the remote resource can be edited. | The specification of the remote resource can be edited. | ||
Revision as of 18:03, 28 February 2006
Contents
Outline
In this tutorial, you will learn how to:
- Create a new Project.
- Load microarray data.
- Merge data from several loaded microarray experiments.
- Rename a project and/or project node.
- Remove a project and/or project node.
- Save project files that you have created.
- Load, add, and/or modify remote data.
Supported data formats
- Microarray
- Affymetrix MAS5/GCOS Files.
- Affymetrix File Matrix - this is the native file type created by geWorkbench.
- RMA Express File - RMA Express is a sophisticated tool for combining data from multiple Affymetrix chips.
- Affy Excel or txt data file.
- Normalized no-confidence expression matrix. A variant of the geWorkbench file matrix format that omits the confidence value columns (P-value or Present/Absent calls).
- Genepix Files - An analysis program for two color arrays.
- Other
- FASTA Files. DNA or protein sequence files in FASTA format.
- Pattern Files.
- Genotypic data Files.
Loading data files into a project
In this example, we will load 10 individual Affymetrix MAS5 format files, and merge them into a single dataset.
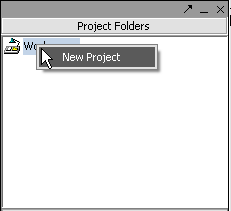
All data must belong to a project. Right-click on the Workspace entry in the Project Folders window at upper left to create a new project.
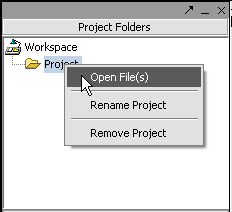
Next, right-click on the New Project entry and select Open Files.
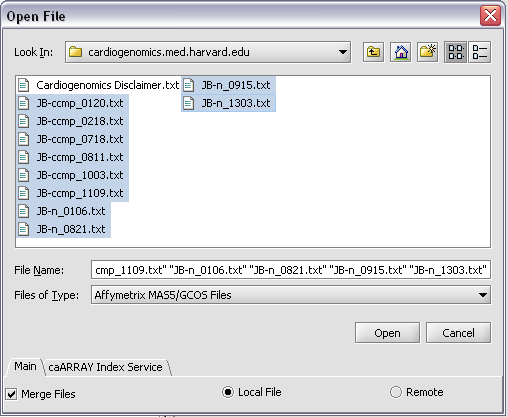
Here, we will select file type Affymetrix GCOS/MAS5 as shown.
Make sure to check the Merge files checkbox.
We select 10 MAS5 format text files from the directory geworkbench\data\training\cardiogenomics.med.harvard.edu, which is included in the geWorkbench download.
Click Open.
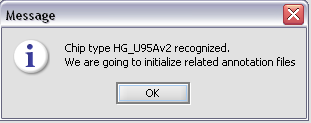
The chip type HG_U95Av2 is recognized...
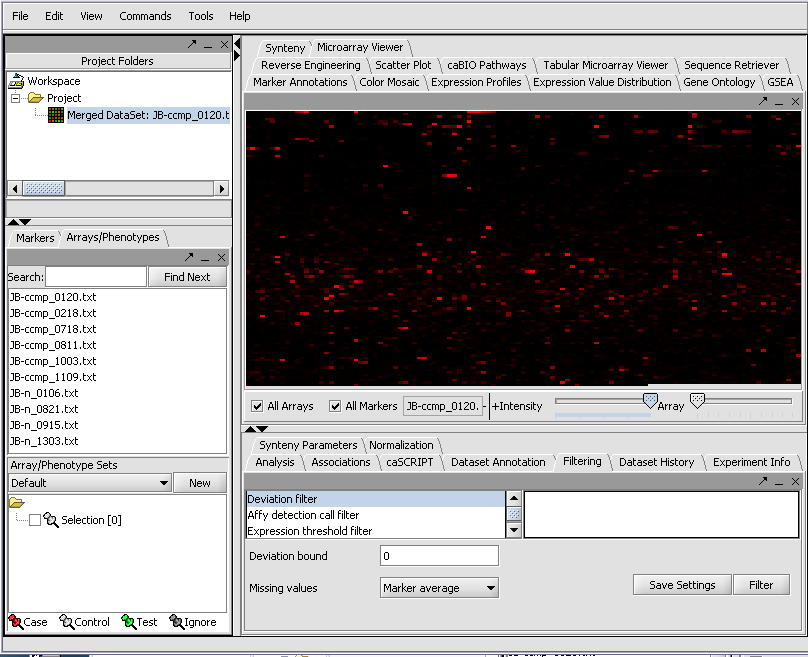
The merged dataset is listed in the Project folder. The data is displayed, in single array format, in the Microarray Viewer. Note we have increased the intensity slider to maximum here.
Merging microarray datafiles after they have already been loaded.
If Affymetrix data files are not merged at the time they are read in, they can also be merged later, as long as they are from the same chip type.
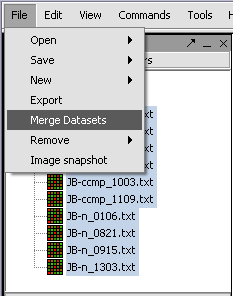
1. Select the read-in data files that you want to merge.
2. Click on File in the menu bar, and choose Merge Datasets.
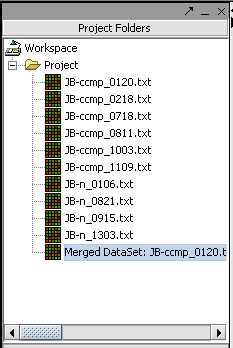
The picture shows the resulting merged dataset created from several individual data files.
The result is a new data node containing the merged data. The original data nodes are still present.
Renaming a project or a data node
Renaming a project
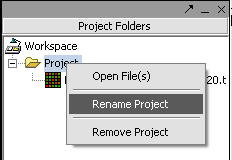
1. Right-click on Project folder.
2. Select Rename.
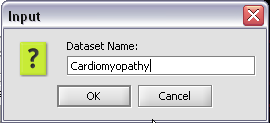
3. In the pop-up screen rename your project.
4. Click on the Okay button
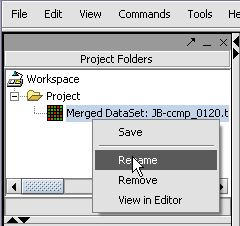
Renaming a project data node
1. Right-click on a Project Folder data node.
2. Select Rename.
3. In the pop-up screen rename your data node.
4. Click on the Okay button.
Removing a project or a data node
Removing a project
1. Right-click on Project folder.
2. Select Remove.
Removing a project data node
1. Right-click on the data node.
2. Select Remove.
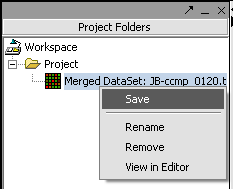
Saving a data node to a file
1. Right-click on data nodes that you want to save.
2. Click Save.
A standard file Save screen will come up.
3. Choose a location.
4. Enter a name.
5. Click on the Save button.
Working with remote data sources
The remote Open File dialog
geWorkbench can retrieve data from certain remote data sources, for example instances of the NCI's caArray database. The Open File dialog allows remote sources to be added to the list of those available either manually or through discovery using grid services. Entries (locations, parameters) for non-grid services can be edited.
As before, right-click on Project which will bring up the Open File dialog. Click the Remote radio button. The Open File dialog window will be expanded to include remote sources.
Four additional buttons appear. They are:
caArray button - Gives you a listing of your Remote Resources.
Go button - Accesses the Remote Source that you selected.
Add A New Resource button - Opens the Data Source Definition Page used to add Remote Data.
Edit button - Edits Remote Source Parameters.
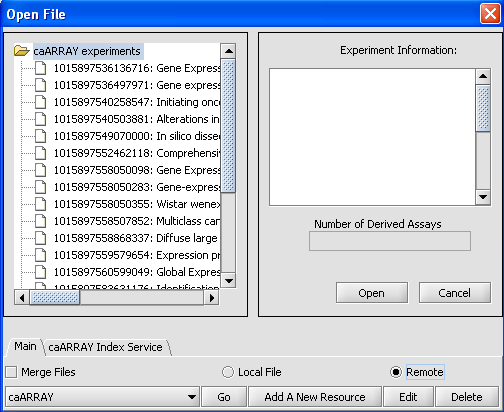
Loading data from a remote instance of caArray
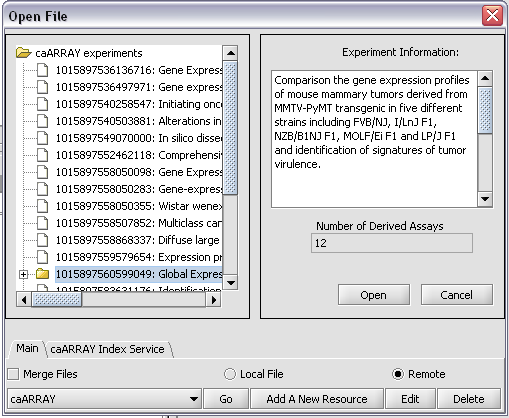
Click on the Go button next to the caArray data source at the bottom of the dialog. All available caArray experiments will be displayed.
Select an experiment that has bioassays. Here we have selected the experiment ending in *99049. The number of derived bioassays, 12, is displayed, along with the experiment information.
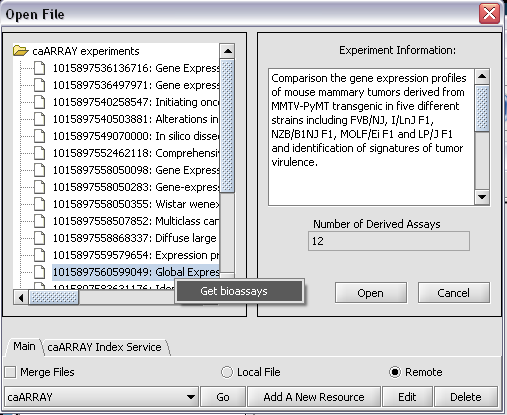
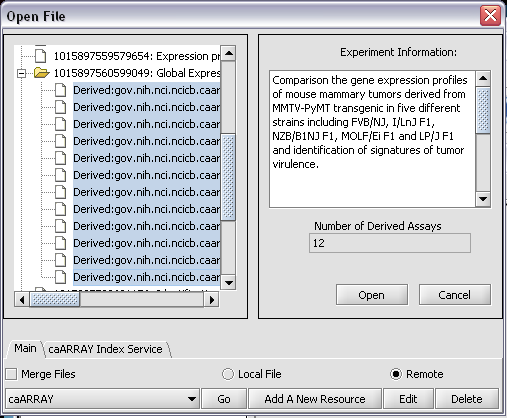
To retrieve the bioassays themselves, right click on the experiment and press Get bioassays. This will download the list of available bioassays into geWorkbench.
To actually retrieve bioassay data, select the desired arrays and push the Open button.
To add a remote source
1. Click on the Add A New Resource button.
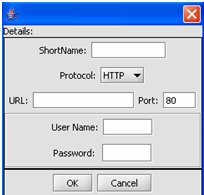 This is the Data Source Definition Page
This is the Data Source Definition Page
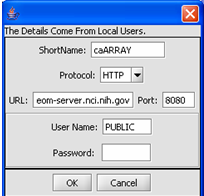
2. Fill in the Data Source definition page. URL and Short Name are required fields.
3. Click on the OK button.
The configuration is set up to automatically reflect your additional Data Source.
To modify a remote source
The specification of the remote resource can be edited.
1. Click on the Edit button.
2. Make the changes that you need.
3. Click on the OK button