Tutorial - Reverse Engineering
Contents
Overview
The Reverse Engineering component included in geWorkbench is being rewritten (as of April 2006), and hence in a future release the details of the interface may change. However, the functionality will be similar.
The primary use of the Reverse Engineering component is to infer regulatory interactions between genes and gene products. The Reverse Engineering component uses the information theory concept of mutual information to find these interactions. Mutual Information is in principle more sensitive and flexible than a simple correlation calculation. It is also invariant under data transformations, so the details of normalization should not be important.
Reverse Engineering in the context of geWorkbench
The Reverse Engineering component calculates the information that the expression pattern of one gene carries about the expression of another gene, that is, it is a pairwise calculation. Larger datasets, containing more arrays per marker, will yield greater sensitivity and better statistical support. Full scale runs of reverse engineering algorithms, comparing all markers against each other, and typically done on datasets containing several hundred microarrays, are typically performed on large cluster computers and are not feasible on a desktop machine. During 2006 we hope to provide a remote service that can host such calculations for jobs launched from geWorkbench. However, at present, smaller scale calculations are supported directly in geWorkbench.
As typically used in geWorkbench, the Reverse Engineering component calculates the Mutual Information score between a single hub gene and all other N markers in the dataset. In a second step, a subset containing the best M markers is chosen (with a current limit of 100), and a complete pairwise MxM/2 mutual information calculation is performed between them. The network resulting from this calculation can be displayed as a branched tree of interactions within the Cytoscape component.
Prerequisites
A dataset containing multiple arrays (the more the better) should be loaded into geWorkbench. If data is loaded from separate files, it should be merged into a single microarray datset, either at the time of or after being read in. See the section Projects and Data Files. In this tutorial we will load a dataset also used in other tutorial sections, which has been normalized, and filtered to reduce the number of genes. This file, "webmatrix_quantile_log2_dev1_mv0.exp" will be made available in the tutorial data section. It can also be obtained by starting with the file "webmatrix.exp" available in the downloads section and performing the following steps:
1. Load the file webmatrix.exp.
2. Quantile Normalize.
3. Log2 transform (also in the Normalize tab).
4. Filter out values having deviation less than 1.
5. Remove markers with filtered-out values using the missing values filter with a threshold of 0.
Example - Profiler
- Load the data file "webmatrix_quantile_log2_dev1_mv0.exp". This contains a set of 100 experiments on Affymetrix HG_U95Av2 chips. As described above, this file has been quantile normalized, log2 converted, and then filtered to remove markers with a deviation of less than 1, in order to reduce the size of the dataset, leaving 3837 markers.
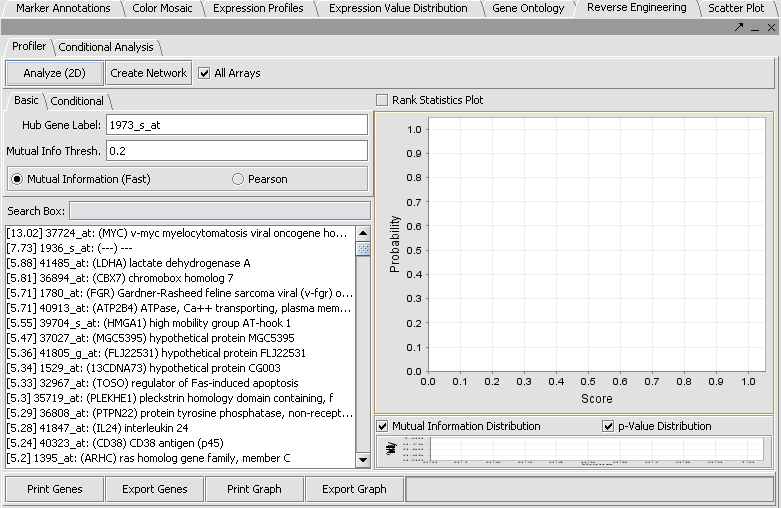
- In the upper right section of geWorkbench find the Reverse Engineering component. It should by default be displaying the Profiler tab.
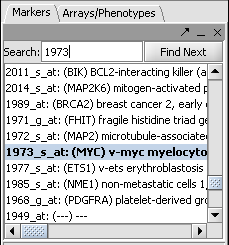
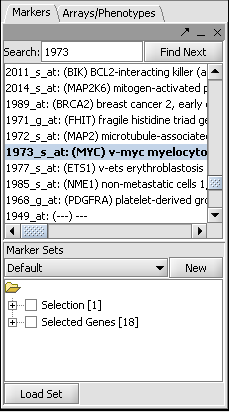
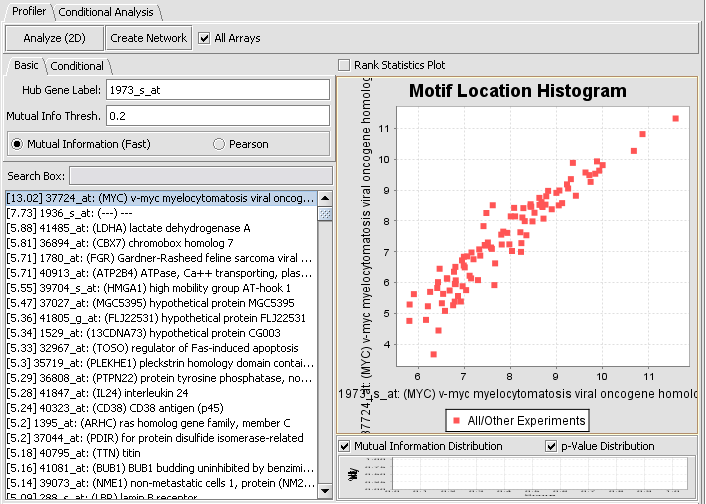
- In the Markers component search box, on the left side of the geWorkbench interface, enter 1973 and hit enter. This will find the marker 1973_s_at, which is the c-Myc gene, a well-known transcription factor with many interactions. Click on this marker in the list. This will enter the marker into the Hub Gene Label field of the Profiler.
- The default setting in the Profiler is Mutual Information (fast). With this selected, hit Analyze(2D). This will return a list of all markers having a MI score of greater than the cutoff value (the default is 0.2).
After the Mutual Information algoritnm has been run, an adjacency matrix will be placed in the Projects Folder:
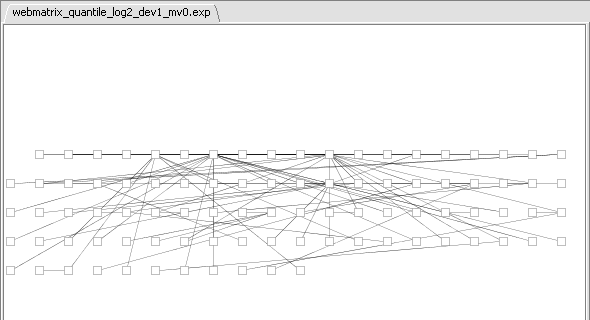
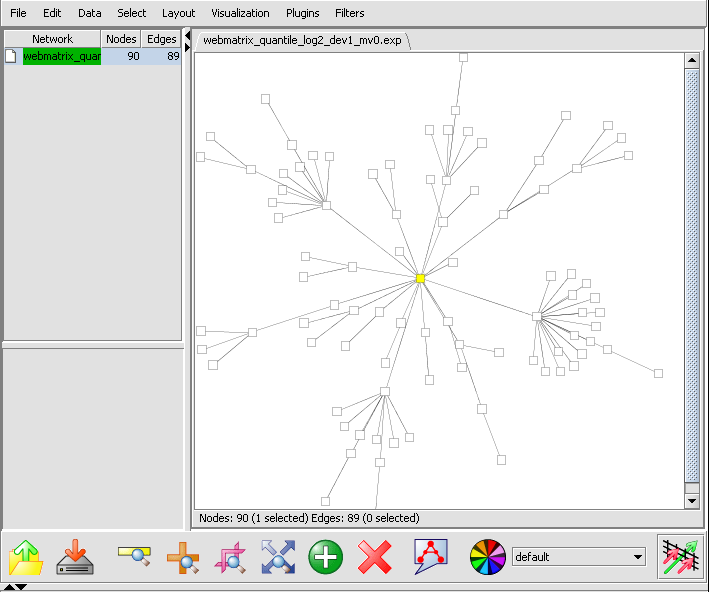
- If at this point you hit the Create Network button, a network will be displayed based on the top 100 markers interacting with c-Myc. As described above, the MI algoritm is run again on these M=100 markers, in order to measure interactions between each pair. Each marker is then connected via an edge with the marker it most strongly interacts with, with the chosen hub-gene at the center.
This is best seen in Cytoscape by going to the Layout menu, and chosing yFiles->organic. The layout will now appear as:
- If a smaller list is desired, a set of markers can be highlighted in the list originally returned. Only this selected subset, up to 100 markers, will then be used if "Create Network" is pressed.
- By right-clicking and selecting "Add to Set", this group will be added to the Markers component as a new set of markers which can be used in other components (sequence retrieval, annotation retriever etc.).
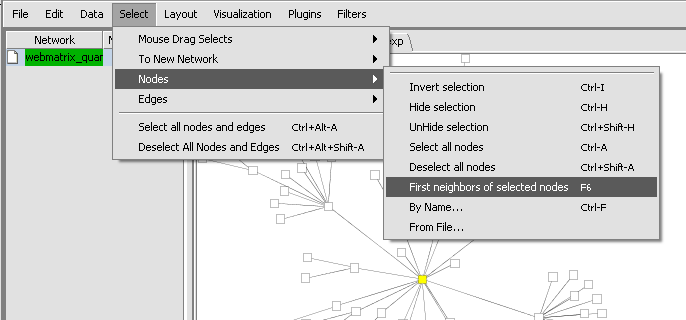
- Within the network created in Cytoscape, one can select the central gene, and then on the Cytoscape menu chose Select->Nodes->First Neighbors of selected nodes.
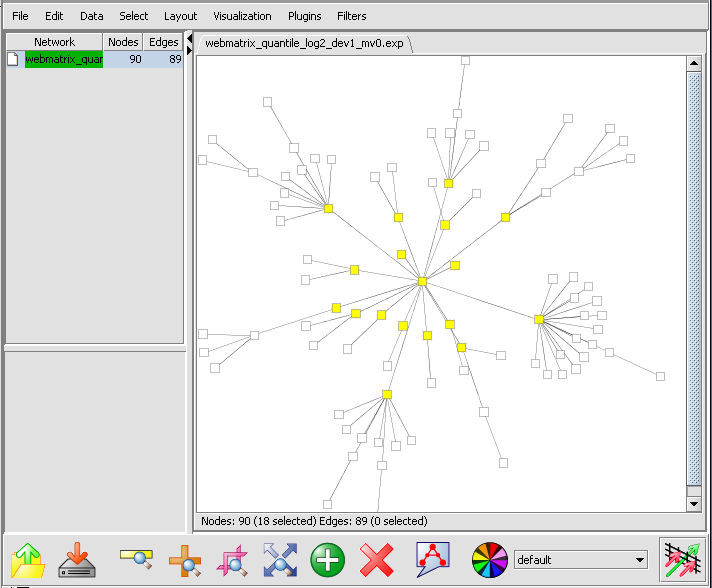
The first neighbors will be highlighted in the graph,
and also added as a new set in the Markers component.
We can return to the main Reverse Engineering component by clicking on the original dataset in the Project Folders component. If we select the first (highest MI score) marker on the list, the graph shown below is drawn in the Motif Location Histogram display. This shows a plot of the expression values on each array for the selected hub marker vs any other marker selected in the list.
Options
Pearson - Uses a Pearson correlation function to calculate the interaction scores.