Difference between revisions of "SOM"
(→Background) |
|||
| (33 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
__TOC__ | __TOC__ | ||
| − | |||
| − | |||
| − | == | + | ==Overview== |
| − | In | + | The SOM (Self-Organizing Maps) method clusters markers into a user-specified number of bins based on their similarity to each other. In geWorkbench, a SOM visualizer component displays the results graphically. The markers clustered into a particular bin can be saved as a set back to the Markers component for further analysis. |
| − | |||
| − | |||
| − | |||
| − | |||
| + | * Note - for the distance calculations used in SOM analysis to be valid, the data should be normalized such that the scale of variation over each array is equal. | ||
| − | + | A more formal description - Self Organizing Map (SOM) is an algorithm to perform clustering of real vectors defined on an instance space of dimensionality. The clusters found are described by prototypical instances, referred as neurons of the SOM, which are arranged topologically in the form of a one or two dimensional grid, the Self Organizing Map. | |
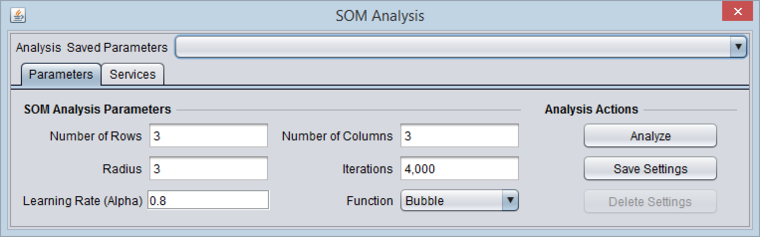
| − | + | ==SOM Parameters== | |
| − | + | [[Image:SOM_Parameters.png|{{ImageMaxWidth}}]] | |
| − | |||
| − | |||
| − | |||
| − | + | ===Number of Rows and Columns=== | |
| + | The final graphical display will be laid out using the given number of rows and columns. The product of the two gives the number of bins into which the markers will be clustered. Both rows and columns must be greater than 0. | ||
| + | |||
| + | ===Radius=== | ||
| + | When using the bubble neighborhood parameter this float value is used to define the extent of the neighborhood. If an SOM vector is within this distance from the winning node (the cluster to which an element has been assigned) then that Node (and SOM vector) is considered to be in the neighborhood and its SOM vector is adapted. This must be a number greater than 0. | ||
| + | |||
| + | ===Iterations=== | ||
| + | The number of times the dataset will be presented to the Map. Each expression element will be presented this number of times to train the Nodes. This must be a number be greater than 0. | ||
| + | |||
| + | |||
| + | ===Alpha=== | ||
| + | This value is used to scale the change of individual SOM vectors when a new expression vector is associated with a node. This must be a value between 1 and 0. | ||
| + | |||
| + | ===Function=== | ||
| + | The neighborhood options indicate the conventions (formulas) used to update (adapt) an SOM vector once an expression vector has been added into a Node's neighborhood. | ||
| + | ====Bubble==== | ||
| + | This option uses the provided radius (see above) to determine which surrounding SOM nodes are in the neighborhood and therefore are candidates for adaptation. When this option is selected the Alpha parameter for scaling the adaptation is used directly as provided from the user. | ||
| + | ====Gaussian==== | ||
| + | This option forces all SOM vectors in the network to be adapted regardless of proximity to the winning node. In this case the Alpha parameter is scaled based on the distance between the SOM vector to be adapted and the winning node's SOM vector. | ||
| + | |||
| + | ==Services (Grid)== | ||
| + | |||
| + | SOM can be run either locally within geWorkbench, or remotely as a grid job on caGrid. See the [[Tutorial_-_Grid_Services | Grid Services]] section for further details on setting up a grid job. | ||
| + | |||
| + | ==Analysis Actions== | ||
| + | This component uses the standard [[Analysis_Framework|analysis component framework]], which provides three buttons: | ||
| + | |||
| + | * '''Analyze''' - Start the clustering job. | ||
| + | * '''Save Settings''' - Save the current settings to a named entry in the settings list. | ||
| + | * '''Delete Settings''' - Delete the selected setting entry from the list. | ||
==SOM Example== | ==SOM Example== | ||
| − | + | For this example we will start with the data set used in the [[Tutorial_-_ANOVA | ANOVA]] tutorial. Briefly, the Bcell-100.exp dataset was quantile normalized and log2 transformed. A set of markers found by ANOVA analysis of four groups of arrays was saved to the Markers component. It is this set of markers which will here be clustered. | |
| − | + | ||
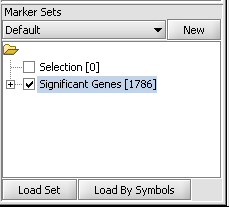
| − | + | 1. If desired, select a subset of markers and/or arrays on which to cluster. The following figure shows the set of 1786 markers found in the ANOVA example has been activated by checking the adjacent check-box. | |
| − | + | ||
| + | [[Image:T_HC_set_activation.png]] | ||
| + | |||
| − | + | 2. Set the SOM parameters. The default parameters are shown above in the Parameters section. We will accept these parameters. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | 3. Push Analyze. The result will be displayed graphically in the SOM Clusters Viewer. | |
| − | + | 4. The user might experiment with different parameter settings to attempt to discern informative groupings. | |
| + | ==SOM Clusters Viewer== | ||
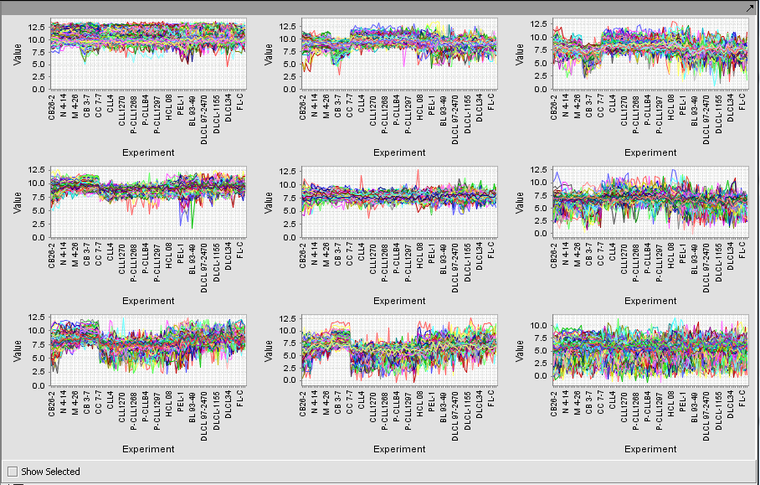
| − | + | Running the example just described, with 3 rows and 3 columns, produces the nine clusters shown below. | |
| − | [[Image: | + | [[Image:T_SOM_result.png|{{ImageMaxWidth}}]] |
| + | ===Mouse over=== | ||
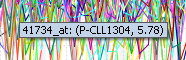
| − | + | Mousing over a particular point on any cluster will show the marker (probeset), the array and the expression value that the point is associated with. | |
| − | + | [[Image:T_SOM_mouse-over.png]] | |
| − | + | ===Show selected=== | |
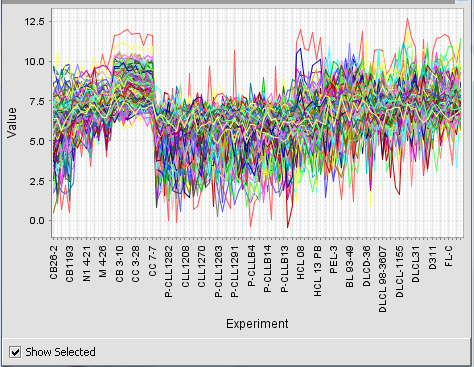
| − | + | If the "Show selected" box is checked, the user may then click on any of the clusters and it will be enlarged to fill the display area. Unchecking the "Show selected" box will return to the original display of all clusters. | |
| − | |||
| − | |||
| − | |||
| + | [[Image:T_SOM_show_selected.png]] | ||
| − | + | ===Left-click actions=== | |
| − | + | ====Selecting==== | |
| − | + | When a point on a cluster is clicked on with the mouse, the marker and array it corresponds to will be highlighted in the Markers and Arrays/Phenotypes components, respectively. | |
| − | |||
| − | + | ====Zooming==== | |
| + | While left-clicking on a cluster display, dragging the mouse downwards and to one side or the other will produce a selection box, which will have the effect of zooming in on the region selected. Dragging the mouse upwards will zoom back out. | ||
| − | + | ===Right-click menu=== | |
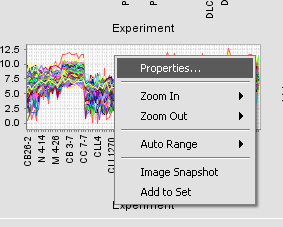
| − | + | Right-clicking on a cluster produces the menu shown below. | |
| − | + | [[Image:T_SOM_right_click_menu.png]] | |
| − | + | The individual choices are: | |
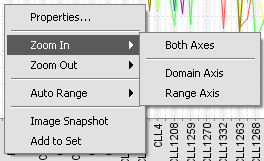
| − | + | ====Zoom In/Zoom Out==== | |
| + | Zoom in or zoom out in a particular cluster. Zooming can be done for both axes simultaneously, or individually using the sub-menus shown in the following figure. | ||
| − | [[Image: | + | [[Image:T_SOM_ZoomIn_detail.png]] |
| + | ====Auto Range==== | ||
| + | Return the cluster to original display size (fit to display area). | ||
| − | + | ====Image Snapshot==== | |
| + | Add a snapshot of the selected cluster to the [[Workspace]]. | ||
| + | ====Add to Set==== | ||
| + | Add the markers in the selected cluster to a new set in the Markers component. The new sets name will start with "Cluster Grid". | ||
| − | |||
| + | ====Properties==== | ||
| + | The "Properties" item allows the title, scale, axis labels and other aspects of the cluster graphs to be customized. | ||
| − | [[Image: | + | [[Image:T_SOM_Properties-title.png]] |
| + | [[Image:T_SOM_Properties-Plot-Domain.png]] | ||
| − | + | [[Image:T_SOM_Properties-Plot-Appearance.png]] | |
| − | [[Image: | + | [[Image:T_SOM_Properties-Other.png]] |
Latest revision as of 19:28, 22 January 2014
Contents
Overview
The SOM (Self-Organizing Maps) method clusters markers into a user-specified number of bins based on their similarity to each other. In geWorkbench, a SOM visualizer component displays the results graphically. The markers clustered into a particular bin can be saved as a set back to the Markers component for further analysis.
- Note - for the distance calculations used in SOM analysis to be valid, the data should be normalized such that the scale of variation over each array is equal.
A more formal description - Self Organizing Map (SOM) is an algorithm to perform clustering of real vectors defined on an instance space of dimensionality. The clusters found are described by prototypical instances, referred as neurons of the SOM, which are arranged topologically in the form of a one or two dimensional grid, the Self Organizing Map.
SOM Parameters
Number of Rows and Columns
The final graphical display will be laid out using the given number of rows and columns. The product of the two gives the number of bins into which the markers will be clustered. Both rows and columns must be greater than 0.
Radius
When using the bubble neighborhood parameter this float value is used to define the extent of the neighborhood. If an SOM vector is within this distance from the winning node (the cluster to which an element has been assigned) then that Node (and SOM vector) is considered to be in the neighborhood and its SOM vector is adapted. This must be a number greater than 0.
Iterations
The number of times the dataset will be presented to the Map. Each expression element will be presented this number of times to train the Nodes. This must be a number be greater than 0.
Alpha
This value is used to scale the change of individual SOM vectors when a new expression vector is associated with a node. This must be a value between 1 and 0.
Function
The neighborhood options indicate the conventions (formulas) used to update (adapt) an SOM vector once an expression vector has been added into a Node's neighborhood.
Bubble
This option uses the provided radius (see above) to determine which surrounding SOM nodes are in the neighborhood and therefore are candidates for adaptation. When this option is selected the Alpha parameter for scaling the adaptation is used directly as provided from the user.
Gaussian
This option forces all SOM vectors in the network to be adapted regardless of proximity to the winning node. In this case the Alpha parameter is scaled based on the distance between the SOM vector to be adapted and the winning node's SOM vector.
Services (Grid)
SOM can be run either locally within geWorkbench, or remotely as a grid job on caGrid. See the Grid Services section for further details on setting up a grid job.
Analysis Actions
This component uses the standard analysis component framework, which provides three buttons:
- Analyze - Start the clustering job.
- Save Settings - Save the current settings to a named entry in the settings list.
- Delete Settings - Delete the selected setting entry from the list.
SOM Example
For this example we will start with the data set used in the ANOVA tutorial. Briefly, the Bcell-100.exp dataset was quantile normalized and log2 transformed. A set of markers found by ANOVA analysis of four groups of arrays was saved to the Markers component. It is this set of markers which will here be clustered.
1. If desired, select a subset of markers and/or arrays on which to cluster. The following figure shows the set of 1786 markers found in the ANOVA example has been activated by checking the adjacent check-box.
2. Set the SOM parameters. The default parameters are shown above in the Parameters section. We will accept these parameters.
3. Push Analyze. The result will be displayed graphically in the SOM Clusters Viewer.
4. The user might experiment with different parameter settings to attempt to discern informative groupings.
SOM Clusters Viewer
Running the example just described, with 3 rows and 3 columns, produces the nine clusters shown below.
Mouse over
Mousing over a particular point on any cluster will show the marker (probeset), the array and the expression value that the point is associated with.
Show selected
If the "Show selected" box is checked, the user may then click on any of the clusters and it will be enlarged to fill the display area. Unchecking the "Show selected" box will return to the original display of all clusters.
Left-click actions
Selecting
When a point on a cluster is clicked on with the mouse, the marker and array it corresponds to will be highlighted in the Markers and Arrays/Phenotypes components, respectively.
Zooming
While left-clicking on a cluster display, dragging the mouse downwards and to one side or the other will produce a selection box, which will have the effect of zooming in on the region selected. Dragging the mouse upwards will zoom back out.
Right-clicking on a cluster produces the menu shown below.
The individual choices are:
Zoom In/Zoom Out
Zoom in or zoom out in a particular cluster. Zooming can be done for both axes simultaneously, or individually using the sub-menus shown in the following figure.
Auto Range
Return the cluster to original display size (fit to display area).
Image Snapshot
Add a snapshot of the selected cluster to the Workspace.
Add to Set
Add the markers in the selected cluster to a new set in the Markers component. The new sets name will start with "Cluster Grid".
Properties
The "Properties" item allows the title, scale, axis labels and other aspects of the cluster graphs to be customized.